A Novel Bacteriophage Exclusion (BREX) System Encoded by the pglX Gene in Lactobacillus casei Zhang
- PMID: 31399407
- PMCID: PMC6805096
- DOI: 10.1128/AEM.01001-19
A Novel Bacteriophage Exclusion (BREX) System Encoded by the pglX Gene in Lactobacillus casei Zhang
Abstract
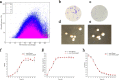
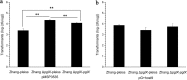
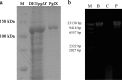
The bacteriophage exclusion (BREX) system is a novel prokaryotic defense system against bacteriophages. To our knowledge, no study has systematically characterized the function of the BREX system in lactic acid bacteria. Lactobacillus casei Zhang is a probiotic bacterium originating from koumiss. By using single-molecule real-time sequencing, we previously identified N6-methyladenine (m6A) signatures in the genome of L. casei Zhang and a putative methyltransferase (MTase), namely, pglX This work further analyzed the genomic locus near the pglX gene and identified it as a component of the BREX system. To decipher the biological role of pglX, an L. casei Zhang pglX mutant (ΔpglX) was constructed. Interestingly, m6A methylation of the 5'-ACRCAG-3' motif was eliminated in the ΔpglX mutant. The wild-type and mutant strains exhibited no significant difference in morphology or growth performance in de Man-Rogosa-Sharpe (MRS) medium. A significantly higher plasmid acquisition capacity was observed for the ΔpglX mutant than for the wild type if the transformed plasmids contained pglX recognition sites (i.e., 5'-ACRCAG-3'). In contrast, no significant difference was observed in plasmid transformation efficiency between the two strains when plasmids lacking pglX recognition sites were tested. Moreover, the ΔpglX mutant had a lower capacity to retain the plasmids than the wild type, suggesting a decrease in genetic stability. Since the Rebase database predicted that the L. casei PglX protein was bifunctional, as both an MTase and a restriction endonuclease, the PglX protein was heterologously expressed and purified but failed to show restriction endonuclease activity. Taken together, the results show that the L. casei Zhang pglX gene is a functional adenine MTase that belongs to the BREX system.IMPORTANCELactobacillus casei Zhang is a probiotic that confers beneficial effects on the host, and it is thus increasingly used in the dairy industry. The possession of an effective bacterial immune system that can defend against invasion of phages and exogenous DNA is a desirable feature for industrial bacterial strains. The bacteriophage exclusion (BREX) system is a recently described phage resistance system in prokaryotes. This work confirmed the function of the BREX system in L. casei and that the methyltransferase (pglX) is an indispensable part of the system. Overall, our study characterizes a BREX system component gene in lactic acid bacteria.
Keywords: BREX; Lactobacillus casei Zhang; MTase; bacteriophage exclusion system; methyltransferase; pglX.
Copyright © 2019 American Society for Microbiology.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

