Intercalating TOP2 Poisons Attenuate Topoisomerase Action at Higher Concentrations
- PMID: 31399497
- PMCID: PMC6744389
- DOI: 10.1124/mol.119.117259
Intercalating TOP2 Poisons Attenuate Topoisomerase Action at Higher Concentrations
Abstract
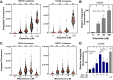
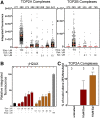
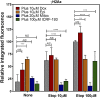
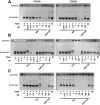
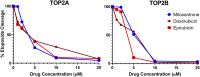
Topoisomerase II (TOP2) poisons are effective cytotoxic anticancer agents that stabilize the normally transient TOP2-DNA covalent complexes formed during the enzyme reaction cycle. These drugs include etoposide, mitoxantrone, and the anthracyclines doxorubicin and epirubicin. Anthracyclines also exert cell-killing activity via TOP2-independent mechanisms, including DNA adduct formation, redox activity, and lipid peroxidation. Here, we show that anthracyclines and another intercalating TOP2 poison, mitoxantrone, stabilize TOP2-DNA covalent complexes less efficiently than etoposide, and at higher concentrations they suppress the formation of TOP2-DNA covalent complexes, thus behaving as TOP2 poisons at low concentration and inhibitors at high concentration. We used induced pluripotent stem cell (iPSC)-derived human cardiomyocytes as a model to study anthracycline-induced damage in cardiac cells. Using immunofluorescence, our study is the first to demonstrate the presence of topoisomerase IIβ (TOP2B) as the only TOP2 isoform in iPSC-derived cardiomyocytes. In these cells, etoposide robustly induced TOP2B covalent complexes, but we could not detect doxorubicin-induced TOP2-DNA complexes, and doxorubicin suppressed etoposide-induced TOP2-DNA complexes. In vitro, etoposide-stabilized DNA cleavage was attenuated by doxorubicin, epirubicin, or mitoxantrone. Clinical use of anthracyclines is associated with cardiotoxicity. The observations in this study have potentially important clinical consequences regarding the effectiveness of anticancer treatment regimens when TOP2-targeting drugs are used in combination. These observations suggest that inhibition of TOP2B activity, rather than DNA damage resulting from TOP2 poisoning, may play a role in doxorubicin cardiotoxicity. SIGNIFICANCE STATEMENT: We show that anthracyclines and mitoxantrone act as topoisomerase II (TOP2) poisons at low concentration but attenuate TOP2 activity at higher concentration, both in cells and in in vitro cleavage experiments. Inhibition of type II topoisomerases suppresses the action of other drugs that poison TOP2. Thus, combinations containing anthracyclines or mitoxantrone and etoposide may reduce the activity of etoposide as a TOP2 poison and thus reduce the efficacy of drug combinations.
Copyright © 2019 by The American Society for Pharmacology and Experimental Therapeutics.
Figures



References
-
- Austin CA, Barot HA, Margerrison EE, Turcatti G, Wingfield P, Hayes MV, Fisher LM. (1990) Structure and partial amino acid sequence of calf thymus DNA topoisomerase II: comparison with other type II enzymes. Biochem Biophys Res Commun 170:763–768. - PubMed
-
- Binaschi M, Farinosi R, Austin CA, Fisher LM, Zunino F, Capranico G. (1998) Human DNA topoisomerase IIalpha-dependent DNA cleavage and yeast cell killing by anthracycline analogues. Cancer Res 58:1886–1892. - PubMed
-
- Bodley A, Liu LF, Israel M, Seshadri R, Koseki Y, Giuliani FC, Kirschenbaum S, Silber R, Potmesil M. (1989) DNA topoisomerase II-mediated interaction of doxorubicin and daunorubicin congeners with DNA. Cancer Res 49:5969–5978. - PubMed
-
- Capranico G, De Isabella P, Tinelli S, Bigioni M, Zunino F. (1993) Similar sequence specificity of mitoxantrone and VM-26 stimulation of in vitro DNA cleavage by mammalian DNA topoisomerase II. Biochemistry 32:3038–3046. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

