An acquired high-risk chromosome instability phenotype in multiple myeloma: Jumping 1q Syndrome
- PMID: 31399558
- PMCID: PMC6689064
- DOI: 10.1038/s41408-019-0226-4
An acquired high-risk chromosome instability phenotype in multiple myeloma: Jumping 1q Syndrome
Abstract
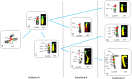
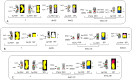
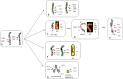
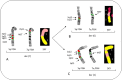
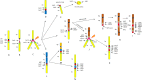
Patients with multiple myeloma (MM) accumulate adverse copy number aberrations (CNAs), gains of 1q21, and 17p deletions during disease progression. A subset of these patients develops heightened 1q12 pericentromeric instability and jumping translocations of 1q12 (JT1q12), evidenced by increased copy CNAs of 1q21 and losses in receptor chromosomes (RC). To understand the progression of these aberrations we analyzed metaphase cells of 50 patients with ≥4 CNAs of 1q21 by G-banding, locus specific FISH, and spectral karyotyping. In eight patients with ≥5 CNAs of 1q21 we identified a chromosome instability phenotype similar to that found in ICF syndrome (immunodeficiency, centromeric instability, and facial anomalies). Strikingly, the acquired instability phenotype identified in these patients demonstrates the same transient structural aberrations of 1q12 as those found in ICF syndrome, suggesting similar underlying pathological mechanisms. Four types of clonal aberrations characterize this phenotype including JT1q12s, RC deletions, 1q12-21 breakage-fusion-bridge cycle amplifications, and RC insertions. In addition, recurring transient aberrations include 1q12 decondensation and breakage, triradials, and 1q micronuclei. The acquired self-propagating mobile property of 1q12 satellite DNA drives the continuous regeneration of 1q12 duplication/deletion events. For patients demonstrating this instability phenotype, we propose the term "Jumping 1q Syndrome."
Conflict of interest statement
The authors declare no competing financial interests.
Figures

References
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

