Rhinovirus-16 induced temporal interferon responses in nasal epithelium links with viral clearance and symptoms
- PMID: 31400236
- PMCID: PMC6972523
- DOI: 10.1111/cea.13481
Rhinovirus-16 induced temporal interferon responses in nasal epithelium links with viral clearance and symptoms
Abstract
Background: The temporal in vivo response of epithelial cells to a viral challenge and its association with viral clearance and clinical outcomes has been largely unexplored in asthma.
Objective: To determine gene expression profiles over time in nasal epithelial cells (NECs) challenged in vivo with rhinovirus-16 (RV16) and compare to nasal symptoms and viral clearance.
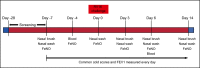
Methods: Patients with stable mild to moderate asthma (n = 20) were challenged intranasally with RV16. Nasal brush samples for RNA sequencing were taken 7 days prior to infection and 3, 6 and 14 days post-infection, and blood samples 4 days prior to infection and day 6 post-infection. Viral load was measured in nasal lavage fluid at day 3, 6 and 14.
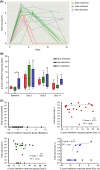
Results: Top differentially (>2.5-fold increase) expressed gene sets in NECs post-RV16 at days 3 and 6, compared with baseline, were interferon alpha and gamma response genes. Patients clearing the virus within 6 days (early resolvers) had a significantly increased interferon response at day 6, whereas those having cleared the virus by day 14 (late resolvers) had significantly increased responses at day 3, 6 and 14. Interestingly, patients not having cleared the virus by day 14 (non-resolvers) had no enhanced interferon responses at any of these days. The daily Cold Symptom Scores (CSS) peaked at days 3 to 5 and correlated positively with interferon response genes at day 3 (R = 0.48), but not at other time-points. Interferon response genes were also enhanced in blood at day 6 after RV16 challenge.
Conclusion and clinical relevance: This study shows that viral load and clearance varies markedly over time in mild to moderate asthma patients exposed to a fixed RV16 dose. The host's nasal interferon response to RV16 at day 3 is associated with upper respiratory tract symptoms. The temporal interferon response in nasal epithelium associates with viral clearance in the nasal compartment.
Keywords: asthma; epithelium; innate immunity; interferon response; nasal epithelium; rhinovirus; virus.
© 2019 The Authors. Clinical & Experimental Allergy published by John Wiley & Sons Ltd.
Conflict of interest statement
The research leading to these results has received support from the Innovative Medicines Initiative (IMI) Joint Undertaking, under grant agreement no. 115010, resources for which are composed of financial contribution from the European Union's Seventh Framework Programme (FP7/2007‐2013) and kind contributions from companies in the European Federation of Pharmaceutical Industries and Associations (EFPIA) (
Figures


References
-
- van der Sluijs KF, van de Pol MA, Kulik W, et al. Systemic tryptophan and kynurenine catabolite levels relate to severity of rhinovirus‐induced asthma exacerbation: a prospective study with a parallel‐group design. Thorax. 2013;68(12):1122‐1130. - PubMed
-
- Corne JM, Marshall C, Smith S, et al. Frequency, severity, and duration of rhinovirus infections in asthmatic and non‐asthmatic individuals: a longitudinal cohort study. Lancet. 2002;359(9309):831‐834. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

