A Pliable Mediator Acts as a Functional Rather Than an Architectural Bridge between Promoters and Enhancers
- PMID: 31402173
- PMCID: PMC7533040
- DOI: 10.1016/j.cell.2019.07.011
A Pliable Mediator Acts as a Functional Rather Than an Architectural Bridge between Promoters and Enhancers
Abstract
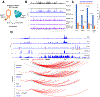
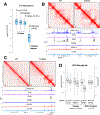
While Mediator plays a key role in eukaryotic transcription, little is known about its mechanism of action. This study combines CRISPR-Cas9 genetic screens, degron assays, Hi-C, and cryoelectron microscopy (cryo-EM) to dissect the function and structure of mammalian Mediator (mMED). Deletion analyses in B, T, and embryonic stem cells (ESC) identified a core of essential subunits required for Pol II recruitment genome-wide. Conversely, loss of non-essential subunits mostly affects promoters linked to multiple enhancers. Contrary to current models, however, mMED and Pol II are dispensable to physically tether regulatory DNA, a topological activity requiring architectural proteins. Cryo-EM analysis revealed a conserved core, with non-essential subunits increasing structural complexity of the tail module, a primary transcription factor target. Changes in tail structure markedly increase Pol II and kinase module interactions. We propose that Mediator's structural pliability enables it to integrate and transmit regulatory signals and act as a functional, rather than an architectural bridge, between promoters and enhancers.
Published by Elsevier Inc.
Conflict of interest statement
DECLARATION OF INTERESTS
The authors declare no conflicts of interest.
Figures





Comment in
-
Mammalian Mediator as a Functional Link between Enhancers and Promoters.Cell. 2019 Aug 22;178(5):1036-1038. doi: 10.1016/j.cell.2019.07.040. Cell. 2019. PMID: 31442397
References
-
- Asturias FJ, Jiang YW, Myers LC, Gustafsson CM, and Kornberg RD (1999). Conserved structures of mediator and RNA polymerase II holoenzyme. Science 283, 985–987. - PubMed
-
- Benabdallah NS, and Bickmore WA (2015). Regulatory Domains and Their Mechanisms. Cold Spring Harb Symp Quant Biol 80, 45–51. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

