Omics Driven Understanding of the Intestines of Parasitic Nematodes
- PMID: 31402928
- PMCID: PMC6669237
- DOI: 10.3389/fgene.2019.00652
Omics Driven Understanding of the Intestines of Parasitic Nematodes
Abstract
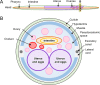
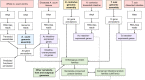
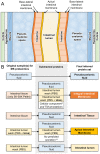
The biological and molecular complexity of nematodes has impeded research on development of new therapies for treatment and control. We have focused on the versatility of the nematode intestine as a target for new therapies. To that end, it is desirable to establish a broad and deep understanding of the molecular architecture underlying intestinal cell functions at the pan-Nematoda level. Multiomics data were generated to uncover the evolutionary principles underlying both conserved and adaptable features of the nematode intestine. Whole genomes were used to reveal the functional potential of the nematodes, tissue-specific transcriptomes provided a deep assessment of genes that are expressed in the adult nematode intestine, and comparison of selected core species was used to determine a first approximation of the pan-Nematoda intestinal transcriptome. Differentially expressed transcripts were also identified among intestinal regions, with the largest number expressed at significantly higher levels in the anterior region, identifying this region as the most functionally unique compared to middle and posterior regions. Profiling intestinal miRNAs targeting these genes identified the conserved intestinal miRNAs. Proteomics of intestinal cell compartments assigned proteins to several different intestinal cell compartments (intestinal tissue, the integral and peripheral intestinal membranes, and the intestinal lumen). Finally, advanced bioinformatic approaches were used to predict intestinal cell functional categories of seminal importance to parasite survival, which can now be experimentally tested and validated. The data provide the most comprehensive compilation of constitutively and differentially expressed genes, predicted gene regulators, and proteins of the nematode intestine. The information provides knowledge that is essential to understand molecular features of nematode intestinal cells and functions of fundamental importance to the intestine of many, if not all, parasitic nematodes.
Keywords: dsRNA; genome; intestine; miRNA; nematode; proteome; transcriptome.
Figures



References
-
- Andrews S. J., Hole N. J., Munn E. A., Rolph T. P. (1995). Vaccination of sheep against haemonchosis with H11, a gut membrane-derived protective antigen from the adult parasite: prevention of the periparturient rise and colostral transfer of protective immunity. Int. J. Parasitol. 25 (7), 839–846. 10.1016/0020-7519(94)00221-9 - DOI - PubMed
-
- Anisimov A. P., Tokmakova N. P., Usheva L. N. (1975). Proliferation and growth of intestinal epithelium in Ascaris suum (Nematoda) during postnatal ontogeny. Communication III. Mitotic anomalies and changes in ploidy and nucleus size. Sov. J. Dev. Biol. 5 (1), 37–44. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

