Conditional Guide RNAs: Programmable Conditional Regulation of CRISPR/Cas Function in Bacterial and Mammalian Cells via Dynamic RNA Nanotechnology
- PMID: 31403072
- PMCID: PMC6661866
- DOI: 10.1021/acscentsci.9b00340
Conditional Guide RNAs: Programmable Conditional Regulation of CRISPR/Cas Function in Bacterial and Mammalian Cells via Dynamic RNA Nanotechnology
Abstract
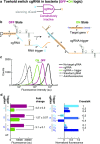
A guide RNA (gRNA) directs the function of a CRISPR protein effector to a target gene of choice, providing a versatile programmable platform for engineering diverse modes of synthetic regulation (edit, silence, induce, bind). However, the fact that gRNAs are constitutively active places limitations on the ability to confine gRNA activity to a desired location and time. To achieve programmable control over the scope of gRNA activity, here we apply principles from dynamic RNA nanotechnology to engineer conditional guide RNAs (cgRNAs) whose activity is dependent on the presence or absence of an RNA trigger. These cgRNAs are programmable at two levels, with the trigger-binding sequence controlling the scope of the effector activity and the target-binding sequence determining the subject of the effector activity. We demonstrate molecular mechanisms for both constitutively active cgRNAs that are conditionally inactivated by an RNA trigger (ON → OFF logic) and constitutively inactive cgRNAs that are conditionally activated by an RNA trigger (OFF → ON logic). For each mechanism, automated sequence design is performed using the reaction pathway designer within NUPACK to design an orthogonal library of three cgRNAs that respond to different RNA triggers. In E. coli expressing cgRNAs, triggers, and silencing dCas9 as the protein effector, we observe a median conditional response of ≈4-fold for an ON → OFF "terminator switch" mechanism, ≈15-fold for an ON → OFF "splinted switch" mechanism, and ≈3-fold for an OFF → ON "toehold switch" mechanism; the median crosstalk within each cgRNA/trigger library is <2%, ≈2%, and ≈20% for the three mechanisms. To test the portability of cgRNA mechanisms prototyped in bacteria to mammalian cells, as well as to test generalizability to different effector functions, we implemented the terminator switch in HEK 293T cells expressing inducing dCas9 as the protein effector, observing a median ON → OFF conditional response of ≈4-fold with median crosstalk of ≈30% for three orthogonal cgRNA/trigger pairs. By providing programmable control over both the scope and target of protein effector function, cgRNA regulators offer a promising platform for synthetic biology.
Conflict of interest statement
The authors declare the following competing financial interest(s): Filed patents.
Figures





References
-
- Gilbert L. A.; Larson M. H.; Morsut L.; Liu Z.; Brar G. A.; Torres S. E.; Stern-Ginossar N.; Brandman O.; Whitehead E. H.; Doudna J. A.; Lim W. A.; Weissman J. S.; Qi L. S. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 2013, 154, 442–451. 10.1016/j.cell.2013.06.044. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

