Translation of Diverse Aramid- and 1,3-Dicarbonyl-peptides by Wild Type Ribosomes in Vitro
- PMID: 31403077
- PMCID: PMC6661870
- DOI: 10.1021/acscentsci.9b00460
Translation of Diverse Aramid- and 1,3-Dicarbonyl-peptides by Wild Type Ribosomes in Vitro
Abstract
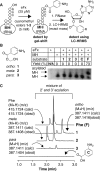
Here, we report that wild type Escherichia coli ribosomes accept and elongate precharged initiator tRNAs acylated with multiple benzoic acids, including aramid precursors, as well as malonyl (1,3-dicarbonyl) substrates to generate a diverse set of aramid-peptide and polyketide-peptide hybrid molecules. This work expands the scope of ribozyme- and ribosome-catalyzed chemical transformations, provides a starting point for in vivo translation engineering efforts, and offers an alternative strategy for the biosynthesis of polyketide-peptide natural products.
Conflict of interest statement
The authors declare no competing financial interest.
Figures




References
-
- Maini R.; Nguyen D. T.; Chen S.; Dedkova L. M.; Chowdhury S. R.; Alcala-Torano R.; Hecht S. M. Incorporation of β-amino acids into dihydrofolate reductase by ribosomes having modifications in the peptidyltransferase center. Bioorg. Med. Chem. 2013, 21, 1088–1096. 10.1016/j.bmc.2013.01.002. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

