Comparative Genomics of Marine Sponge-Derived Streptomyces spp. Isolates SM17 and SM18 With Their Closest Terrestrial Relatives Provides Novel Insights Into Environmental Niche Adaptations and Secondary Metabolite Biosynthesis Potential
- PMID: 31404169
- PMCID: PMC6676996
- DOI: 10.3389/fmicb.2019.01713
Comparative Genomics of Marine Sponge-Derived Streptomyces spp. Isolates SM17 and SM18 With Their Closest Terrestrial Relatives Provides Novel Insights Into Environmental Niche Adaptations and Secondary Metabolite Biosynthesis Potential
Erratum in
-
Corrigendum: Comparative Genomics of Marine Sponge-Derived Streptomyces spp. Isolates SM17 and SM18 With Their Closest Terrestrial Relatives Provides Novel Insights Into Environmental Niche Adaptations and Secondary Metabolite Biosynthesis Potential.Front Microbiol. 2019 Sep 24;10:2213. doi: 10.3389/fmicb.2019.02213. eCollection 2019. Front Microbiol. 2019. PMID: 31595171 Free PMC article.
Abstract
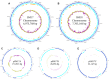
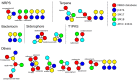
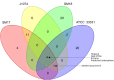
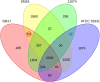
The emergence of antibiotic resistant microorganisms has led to an increased need for the discovery and development of novel antimicrobial compounds. Frequent rediscovery of the same natural products (NPs) continues to decrease the likelihood of the discovery of new compounds from soil bacteria. Thus, efforts have shifted toward investigating microorganisms and their secondary metabolite biosynthesis potential, from diverse niche environments, such as those isolated from marine sponges. Here we investigated at the genomic level two Streptomyces spp. strains, namely SM17 and SM18, isolated from the marine sponge Haliclona simulans, with previously reported antimicrobial activity against clinically relevant pathogens; using single molecule real-time (SMRT) sequencing. We performed a series of comparative genomic analyses on SM17 and SM18 with their closest terrestrial relatives, namely S. albus J1074 and S. pratensis ATCC 33331 respectively; in an effort to provide further insights into potential environmental niche adaptations (ENAs) of marine sponge-associated Streptomyces, and on how these adaptations might be linked to their secondary metabolite biosynthesis potential. Prediction of secondary metabolite biosynthetic gene clusters (smBGCs) indicated that, even though the marine isolates are closely related to their terrestrial counterparts at a genomic level; they potentially produce different compounds. SM17 and SM18 displayed a better ability to grow in high salinity medium when compared to their terrestrial counterparts, and further analysis of their genomes indicated that they possess a pool of 29 potential ENA genes that are absent in S. albus J1074 and S. pratensis ATCC 33331. This ENA gene pool included functional categories of genes that are likely to be related to niche adaptations and which could be grouped based on potential biological functions such as osmotic stress, defense; transcriptional regulation; symbiotic interactions; antimicrobial compound production and resistance; ABC transporters; together with horizontal gene transfer and defense-related features.
Keywords: Streptomyces; comparative genomics; environmental adaptation; marine sponge bacteria; secondary metabolite biosynthetic gene clusters; single molecule real-time sequencing.
Figures



References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

