miRDRN-miRNA disease regulatory network: a tool for exploring disease and tissue-specific microRNA regulatory networks
- PMID: 31404401
- PMCID: PMC6688598
- DOI: 10.7717/peerj.7309
miRDRN-miRNA disease regulatory network: a tool for exploring disease and tissue-specific microRNA regulatory networks
Abstract
Background: MicroRNA (miRNA) regulates cellular processes by acting on specific target genes, and cellular processes proceed through multiple interactions often organized into pathways among genes and gene products. Hundreds of miRNAs and their target genes have been identified, as are many miRNA-disease associations. These, together with huge amounts of data on gene annotation, biological pathways, and protein-protein interactions are available in public databases. Here, using such data we built a database and web service platform, miRNA disease regulatory network (miRDRN), for users to construct disease and tissue-specific miRNA-protein regulatory networks, with which they may explore disease related molecular and pathway associations, or find new ones, and possibly discover new modes of drug action.
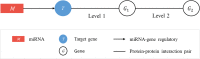
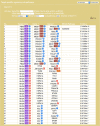
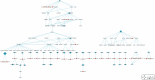
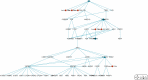
Methods: Data on disease-miRNA association, miRNA-target association and validation, gene-tissue association, gene-tumor association, biological pathways, human protein interaction, gene ID, gene ontology, gene annotation, and product were collected from publicly available databases and integrated. A large set of miRNA target-specific regulatory sub-pathways (RSPs) having the form (T, G 1, G 2) was built from the integrated data and stored, where T is a miRNA-associated target gene, G 1 (G 2) is a gene/protein interacting with T (G 1). Each sequence (T, G 1, G 2) was assigned a p-value weighted by the participation of the three genes in molecular interactions and reaction pathways.
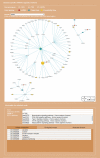
Results: A web service platform, miRDRN (http://mirdrn.ncu.edu.tw/mirdrn/), was built. The database part of miRDRN currently stores 6,973,875 p-valued RSPs associated with 116 diseases in 78 tissue types built from 207 diseases-associated miRNA regulating 389 genes. miRDRN also provides facilities for the user to construct disease and tissue-specific miRNA regulatory networks from RSPs it stores, and to download and/or visualize parts or all of the product. User may use miRDRN to explore a single disease, or a disease-pair to gain insights on comorbidity. As demonstrations, miRDRN was applied: to explore the single disease colorectal cancer (CRC), in which 26 novel potential CRC target genes were identified; to study the comorbidity of the disease-pair Alzheimer's disease-Type 2 diabetes, in which 18 novel potential comorbid genes were identified; and, to explore possible causes that may shed light on recent failures of late-phase trials of anti-AD, BACE1 inhibitor drugs, in which genes downstream to BACE1 whose suppression may affect signal transduction were identified.
Keywords: Alzheimer’s disease; Colorectal cancer; Comorbidity gene; Database and web service tool; Disease and tissue-specific miRNA-protein regulatory network; Disease-miRNA association; Target-specific regulatory pathway; Type 2 diabetes; anti-AD BACE1 inhibitor drug; miRNA-target association.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
-
- Abner EL, Nelson PT, Kryscio RJ, Schmitt FA, Fardo DW, Woltjer RL, Cairns NJ, Yu L, Dodge HH, Xiong C, Masaki K, Tyas SL, Bennett DA, Schneider JA, Arvanitakis Z. Diabetes is associated with cerebrovascular but not Alzheimer neuropathology. Alzheimer’s & Dementia. 2016;12(8):882–889. doi: 10.1016/j.jalz.2015.12.006. - DOI - PMC - PubMed
-
- Ali RH, Marafie MJ, Bitar MS, Al-Dousari F, Ismael S, Bin Haider H, Al-Ali W, Jacob SP, Al-Mulla F. Gender-associated genomic differences in colorectal cancer: clinical insight from feminization of male cancer cells. International Journal of Molecular Sciences. 2014;15(10):17344–17365. doi: 10.3390/ijms151017344. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

