A high-bias, low-variance introduction to Machine Learning for physicists
- PMID: 31404441
- PMCID: PMC6688775
- DOI: 10.1016/j.physrep.2019.03.001
A high-bias, low-variance introduction to Machine Learning for physicists
Abstract
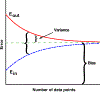
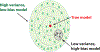
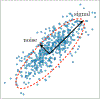
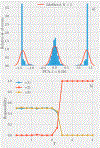
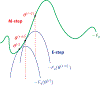
Machine Learning (ML) is one of the most exciting and dynamic areas of modern research and application. The purpose of this review is to provide an introduction to the core concepts and tools of machine learning in a manner easily understood and intuitive to physicists. The review begins by covering fundamental concepts in ML and modern statistics such as the bias-variance tradeoff, overfitting, regularization, generalization, and gradient descent before moving on to more advanced topics in both supervised and unsupervised learning. Topics covered in the review include ensemble models, deep learning and neural networks, clustering and data visualization, energy-based models (including MaxEnt models and Restricted Boltzmann Machines), and variational methods. Throughout, we emphasize the many natural connections between ML and statistical physics. A notable aspect of the review is the use of Python Jupyter notebooks to introduce modern ML/statistical packages to readers using physics-inspired datasets (the Ising Model and Monte-Carlo simulations of supersymmetric decays of proton-proton collisions). We conclude with an extended outlook discussing possible uses of machine learning for furthering our understanding of the physical world as well as open problems in ML where physicists may be able to contribute.
Figures




















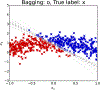
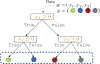





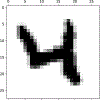





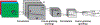

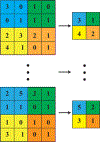
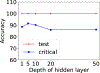

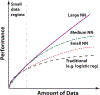




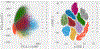














References
-
- Abu-Mostafa, Yaser S, Magdon-Ismail Malik, and Hsuan-Tien Lin (2012), Learning from data, Vol. 4 (AMLBook New York, NY, USA:).
-
- Ackley, David H, Hinton Geoffrey E, and Sejnowski Terrence J (1987), “A learning algorithm for boltzmann machines,” in Readings in Computer Vision (Elsevier; ) pp. 522–533.
-
- Adam, Alison (2006), Artificial knowing: Gender and the thinking machine (Routledge; ).
-
- Advani Madhu, and Ganguli Surya (2016), “Statistical mechanics of optimal convex inference in high dimensions,” Physical Review X 6 (3), 031034.
-
- Advani Madhu, Lahiri Subhaneil, and Ganguli Surya (2013), “Statistical mechanics of complex neural systems and high dimensional data,” Journal of Statistical Mechanics: Theory and Experiment 2013 (03), P03014.
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
