Analysis of Drought Tolerance and Associated Traits in Upland Cotton at the Seedling Stage
- PMID: 31404956
- PMCID: PMC6720584
- DOI: 10.3390/ijms20163888
Analysis of Drought Tolerance and Associated Traits in Upland Cotton at the Seedling Stage
Abstract
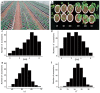
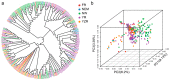
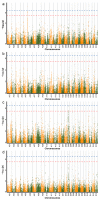
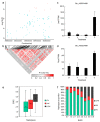
(1) Background: Upland cotton (Gossypium hirsutum L.) is the most important natural fiber worldwide, and it is extensively planted and plentifully used in the textile industry. Major cotton planting regions are frequently affected by abiotic stress, especially drought stress. Drought resistance is a complex, quantitative trait. A genome-wide association study (GWAS) constitutes an efficient method for dissecting the genetic architecture of complex traits. In this study, the drought resistance of a population of 316 upland cotton accessions was studied via GWAS. (2) Methods: GWAS methodology was employed to identify relationships between molecular markers or candidate genes and phenotypes of interest. (3) Results: A total of 8, 3, and 6 SNPs were associated with the euphylla wilting score (EWS), cotyledon wilting score (CWS), and leaf temperature (LT), respectively, based on a general linear model and a factored spectrally transformed linear mixed model. For these traits, 7 QTLs were found, of which 2 each were located on chromosomes A05, A11, and D03, and of which 1 was located on chromosome A01. Importantly, in the candidate regions WRKY70, GhCIPK6, SnRK2.6, and NET1A, which are involved in the response to abscisic acid (ABA), the mitogen-activated protein kinase (MAPK) signaling pathway and the calcium transduction pathway were identified in upland cotton at the seedling stage under drought stress according to annotation information and linkage disequilibrium (LD) block analysis. Moreover, RNA sequencing analysis showed that WRKY70, GhCIPK6, SnRK2.6, and NET1A were induced by drought stress, and the expression of these genes was significantly different between normal and drought stress conditions. (4) Conclusions: The present study should provide some genomic resources for drought resistance in upland cotton. Moreover, the germplasm of the different phenotypes, the detected SNPs and, the potential candidate genes will be helpful for molecular marker-assisted breeding studies about increased drought resistance in upland cotton.
Keywords: association analysis; drought tolerance; single-nucleotide polymorphism (SNP); upland cotton.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
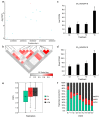
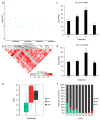
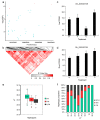
References
-
- Rizza F., Badeck F.W., Cattivelli L., Lidestri O., Di Fonzo N., Stanca A.M. Use of a water stress index to identify barley genotypes adapted to rainfed and irrigated conditions. Crop Sci. 2004;44:2127–2137. doi: 10.2135/cropsci2004.2127. - DOI
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

