Molecular and biological properties of phytoplasmas
- PMID: 31406061
- PMCID: PMC6766451
- DOI: 10.2183/pjab.95.028
Molecular and biological properties of phytoplasmas
Abstract
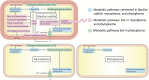
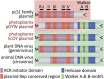
Phytoplasmas, a large group of plant-pathogenic, phloem-inhabiting bacteria were discovered by Japanese scientists in 1967. They are transmitted from plant to plant by phloem-feeding insect hosts and cause a variety of symptoms and considerable damage in more than 1,000 plant species. In the first quarter century following the discovery of phytoplasmas, their tiny cell size and the difficulty in culturing them hampered their biological classification and restricted research to ecological studies such as detection by electron microscopy and identification of insect vectors. In the 1990s, however, tremendous advances in molecular biology and related technologies encouraged investigation of phytoplasmas at the molecular level. In the last quarter century, molecular biology has revealed important properties of phytoplasmas. This review summarizes the history and current status of phytoplasma research, focusing on their discovery, molecular classification, diagnosis of phytoplasma diseases, reductive evolution of their genomes, characteristic features of their plasmids, molecular mechanisms of insect transmission, virulence factors, and chemotherapy.
Keywords: genome; host specificity; mycoplasma-like organism; pathogenicity; phytoplasma; plant pathology.
Figures













Similar articles
-
Phytoplasmas and their interactions with hosts.Trends Plant Sci. 2005 Nov;10(11):526-35. doi: 10.1016/j.tplants.2005.09.008. Epub 2005 Oct 12. Trends Plant Sci. 2005. PMID: 16226054 Review.
-
Phylogenetic positions of 'Candidatus Phytoplasma asteris' and Spiroplasma kunkelii as inferred from multiple sets of concatenated core housekeeping proteins.Int J Syst Evol Microbiol. 2005 Sep;55(Pt 5):2131-2141. doi: 10.1099/ijs.0.63655-0. Int J Syst Evol Microbiol. 2005. PMID: 16166721
-
Phytoplasmas: genetics, diagnosis and relationships with the plant and insect host.Front Biosci. 2007 Jan 1;12:1353-75. doi: 10.2741/2153. Front Biosci. 2007. PMID: 17127387 Review.
-
Phytoplasma: A plant pathogen that cannot be ignored in agricultural production-Research progress and outlook.Mol Plant Pathol. 2024 Feb;25(2):e13437. doi: 10.1111/mpp.13437. Mol Plant Pathol. 2024. PMID: 38393681 Free PMC article. Review.
-
Spiroplasmas and phytoplasmas: microbes associated with plant hosts.Biologicals. 2010 Mar;38(2):193-203. doi: 10.1016/j.biologicals.2009.11.007. Epub 2010 Feb 11. Biologicals. 2010. PMID: 20153217 Review.
Cited by
-
Zinc-Based Nanomaterials for Diagnosis and Management of Plant Diseases: Ecological Safety and Future Prospects.J Fungi (Basel). 2020 Oct 13;6(4):222. doi: 10.3390/jof6040222. J Fungi (Basel). 2020. PMID: 33066193 Free PMC article. Review.
-
Galleria mellonella Invertebrate Model Mirrors the Pathogenic Potential of Mycoplasma alligatoris within the Natural Host.Transbound Emerg Dis. 2024 Mar 22;2024:3009838. doi: 10.1155/2024/3009838. eCollection 2024. Transbound Emerg Dis. 2024. PMID: 40303151 Free PMC article.
-
Impact of the "Flavescence Dorée" Phytoplasma on Xylem Growth and Anatomical Characteristics in Trunks of 'Chardonnay' Grapevines (Vitis vinifera).Biology (Basel). 2022 Jun 28;11(7):978. doi: 10.3390/biology11070978. Biology (Basel). 2022. PMID: 36101359 Free PMC article.
-
Editorial: New insights in small fruit diseases.Front Plant Sci. 2023 Nov 8;14:1306301. doi: 10.3389/fpls.2023.1306301. eCollection 2023. Front Plant Sci. 2023. PMID: 38023924 Free PMC article. No abstract available.
-
A phytoplasma effector acts as a ubiquitin-like mediator between floral MADS-box proteins and proteasome shuttle proteins.Plant Cell. 2022 Apr 26;34(5):1709-1723. doi: 10.1093/plcell/koac062. Plant Cell. 2022. PMID: 35234248 Free PMC article.
References
-
- Wang, M. and Maramorosch, K. (1988) Earliest historical record of a tree mycoplasma disease: Beneficial effect of mycoplasma-like organisms on peonies. In Mycoplasma Diseases of Crops: Basic and Applied Aspects (eds. Maramorosch, K. and Raychaudhuri, S.P.). Springer-Verlag, New York, pp. 349–356.
-
- The Phytopathological Society of Japan (2018) Common Names of Plant Diseases in Japan (2018 edition). The Phytopathological Society of Japan, Tokyo (in Japanese).
-
- Ishiie T. (1965) Dwarf disease of mulberry tree. Ann. Phytopathol. Soc. Jpn. 31, 139–144 (in Japanese).
-
- Strauss E. (2009) Phytoplasma research begins to bloom. Science 325, 388–390. - PubMed
-
- Brown S.E., Been B.O., McLaughlin W.A. (2006) Detection and variability of the lethal yellowing group (16Sr IV) phytoplasmas in the Cedusa sp. (Hemiptera: Auchenorrhyncha: Derbidae) in Jamaica. Ann. Appl. Biol. 149, 53–62.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

