Trichoderma harzianum favours the access of arbuscular mycorrhizal fungi to non-host Brassicaceae roots and increases plant productivity
- PMID: 31406170
- PMCID: PMC6690897
- DOI: 10.1038/s41598-019-48269-z
Trichoderma harzianum favours the access of arbuscular mycorrhizal fungi to non-host Brassicaceae roots and increases plant productivity
Abstract
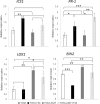
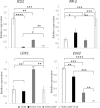
The family Brassicaceae includes plants that are non-host for arbuscular mycorrhizal fungi (AMF) such as the model plant Arabidopsis thaliana (arabidopsis) and the economically important crop plant Brassica napus (rapeseed). It is well known that Trichoderma species have the ability to colonize the rhizosphere of Brassicaceae plants, promoting growth and development as well as stimulating systemic defenses. The aim of the present work is to ascertain that Brassicaceae plants increase productivity when AMF and Trichoderma are combinedly applied, and how such an effect can be ruled. This simultaneous application of a Trichoderma harzianum biocontrol strain and an AMF formulation produces a significant increase in the colonization by Trichoderma and the presence of AMF in arabidopsis and rapeseed roots, such colonization accompanied by improved productivity in both Brassicaceae species. Expression profiling of defense-related marker genes suggests that the phytohormone salicylic acid plays a key role in the modulation of the root colonization process when both fungi are jointly applied.
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Mycorrhizal fungi and Trichoderma harzianum as biocontrol agents for suppression of Rhizoctonia solani damping-off disease of tomato.Commun Agric Appl Biol Sci. 2008;73(2):217-32. Commun Agric Appl Biol Sci. 2008. PMID: 19226759
-
The interaction with arbuscular mycorrhizal fungi or Trichoderma harzianum alters the shoot hormonal profile in melon plants.Phytochemistry. 2011 Feb;72(2-3):223-9. doi: 10.1016/j.phytochem.2010.11.008. Epub 2010 Dec 7. Phytochemistry. 2011. PMID: 21145086
-
Induction of SA-signaling pathway and ethylene biosynthesis in Trichoderma harzianum-treated tomato plants after infection of the root-knot nematode Meloidogyne incognita.Plant Cell Rep. 2017 Apr;36(4):621-631. doi: 10.1007/s00299-017-2109-0. Epub 2017 Feb 26. Plant Cell Rep. 2017. PMID: 28239746
-
Weights in the balance: jasmonic acid and salicylic acid signaling in root-biotroph interactions.Mol Plant Microbe Interact. 2009 Jul;22(7):763-72. doi: 10.1094/MPMI-22-7-0763. Mol Plant Microbe Interact. 2009. PMID: 19522558 Review.
-
The interactions of Trichoderma at multiple trophic levels: inter-kingdom communication.Microbiol Res. 2020 Nov;240:126552. doi: 10.1016/j.micres.2020.126552. Epub 2020 Jul 7. Microbiol Res. 2020. PMID: 32659716 Review.
Cited by
-
When Salt Meddles Between Plant, Soil, and Microorganisms.Front Plant Sci. 2020 Sep 16;11:553087. doi: 10.3389/fpls.2020.553087. eCollection 2020. Front Plant Sci. 2020. PMID: 33042180 Free PMC article. Review.
-
A coumarin exudation pathway mitigates arbuscular mycorrhizal incompatibility in Arabidopsis thaliana.Plant Mol Biol. 2021 Jul;106(4-5):319-334. doi: 10.1007/s11103-021-01143-x. Epub 2021 Apr 6. Plant Mol Biol. 2021. PMID: 33825084
-
Trichoderma Isolates Against Abiotic Stresses and Management of Collar rot of Lentil (Lens culinaris L.) Caused by Sclerotium rolfsii.Indian J Microbiol. 2024 Sep;64(3):1366-1375. doi: 10.1007/s12088-024-01356-w. Epub 2024 Jul 27. Indian J Microbiol. 2024. PMID: 39282175 Free PMC article.
-
The Functional Profile and Antioxidant Capacity of Tomato Fruits Are Modulated by the Interaction between Microbial Biostimulants, Soil Properties, and Soil Nitrogen Status.Antioxidants (Basel). 2023 Feb 19;12(2):520. doi: 10.3390/antiox12020520. Antioxidants (Basel). 2023. PMID: 36830078 Free PMC article.
-
Shifting Perspectives of Translational Research in Bio-Bactericides: Reviewing the Bacillus amyloliquefaciens Paradigm.Biology (Basel). 2021 Nov 18;10(11):1202. doi: 10.3390/biology10111202. Biology (Basel). 2021. PMID: 34827195 Free PMC article. Review.
References
-
- Baum C, El-Tohamy W, Gruda N. Increasing the productivity and product quality of vegetable crops using arbuscular mycorrhizal fungi: a review. Sci. Hortic. 2015;187:131–141. doi: 10.1016/j.scienta.2015.03.002. - DOI
-
- Veresoglou SD, Chen B, Rillig MC. Arbuscular mycorrhiza and soil nitrogen cycling. Soil Biol. Biochem. 2012;46:53–62. doi: 10.1016/j.soilbio.2011.11.018. - DOI
-
- Latef AAHA, et al. Arbuscular mycorrhizal symbiosis and abiotic stress in plants: a review. J. Plant Biol. 2016;59:407–426. doi: 10.1007/s12374-016-0237-7. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

