DABs are inorganic carbon pumps found throughout prokaryotic phyla
- PMID: 31406332
- PMCID: PMC10184468
- DOI: 10.1038/s41564-019-0520-8
DABs are inorganic carbon pumps found throughout prokaryotic phyla
Abstract
Bacterial autotrophs often rely on CO2 concentrating mechanisms (CCMs) to assimilate carbon. Although many CCM proteins have been identified, a systematic screen of the components of CCMs is lacking. Here, we performed a genome-wide barcoded transposon screen to identify essential and CCM-related genes in the γ-proteobacterium Halothiobacillus neapolitanus. Screening revealed that the CCM comprises at least 17 and probably no more than 25 genes, most of which are encoded in 3 operons. Two of these operons (DAB1 and DAB2) contain a two-gene locus that encodes a domain of unknown function (Pfam: PF10070) and a putative cation transporter (Pfam: PF00361). Physiological and biochemical assays demonstrated that these proteins-which we name DabA and DabB, for DABs accumulate bicarbonate-assemble into a heterodimeric complex, which contains a putative β-carbonic anhydrase-like active site and functions as an energy-coupled inorganic carbon (Ci) pump. Interestingly, DAB operons are found in a diverse range of bacteria and archaea. We demonstrate that functional DABs are present in the human pathogens Bacillus anthracis and Vibrio cholerae. On the basis of these results, we propose that DABs constitute a class of energized Ci pumps and play a critical role in the metabolism of Ci throughout prokaryotic phyla.
Conflict of interest statement
Competing Interests
UC Regents have filed a patent related to this work on which J.J.D., A.F., and D.F.S. are inventors. D.F.S. is a co-founder of Scribe Therapeutics and a scientific advisory board member of Scribe Therapeutics and Mammoth Biosciences. All other authors declare no competing interests.
Figures




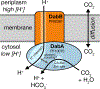
Comment in
-
DABs accumulate bicarbonate.Nat Microbiol. 2019 Dec;4(12):2029-2030. doi: 10.1038/s41564-019-0629-9. Nat Microbiol. 2019. PMID: 31754274 No abstract available.
References
-
- Bathellier C, Tcherkez G, Lorimer GH & Farquhar GD Rubisco is not really so bad. Plant Cell Environ. 41, 705–716 (2018). - PubMed
-
- Flamholz A et al. Revisiting tradeoffs in Rubisco kinetic parameters. bioRxiv 470021 (2018). doi: 10.1101/470021 - DOI
-
- Tcherkez G The mechanism of Rubisco-catalysed oxygenation. Plant Cell Environ. 39, 983–997 (2016). - PubMed
-
- Bauwe H, Hagemann M & Fernie AR Photorespiration: players, partners and origin. Trends Plant Sci. 15, 330–336 (2010). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

