Targeted sequencing reveals expanded genetic diversity of human transfer RNAs
- PMID: 31407949
- PMCID: PMC6779403
- DOI: 10.1080/15476286.2019.1646079
Targeted sequencing reveals expanded genetic diversity of human transfer RNAs
Abstract
Transfer RNAs are required to translate genetic information into proteins as well as regulate other cellular processes. Nucleotide changes in tRNAs can result in loss or gain of function that impact the composition and fidelity of the proteome. Despite links between tRNA variation and disease, the importance of cytoplasmic tRNA variation has been overlooked. Using a custom capture panel, we sequenced 605 human tRNA-encoding genes from 84 individuals. We developed a bioinformatic pipeline that allows more accurate tRNA read mapping and identifies multiple polymorphisms occurring within the same variant. Our analysis identified 522 unique tRNA-encoding sequences that differed from the reference genome from 84 individuals. Each individual had ~66 tRNA variants including nine variants found in less than 5% of our sample group. Variants were identified throughout the tRNA structure with 17% predicted to enhance function. Eighteen anticodon mutants were identified including potentially mistranslating tRNAs; e.g., a tRNASer that decodes Phe codons. Similar engineered tRNA variants were previously shown to inhibit cell growth, increase apoptosis and induce the unfolded protein response in mammalian cell cultures and chick embryos. Our analysis shows that human tRNA variation has been underestimated. We conclude that the large number of tRNA genes provides a buffer enabling the emergence of variants, some of which could contribute to disease.
Keywords: human tRNA variation; tRNA biology; tRNA capture panel; tRNA-encoding genes; translation.
Figures
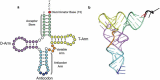

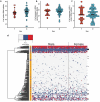




Similar articles
-
Actions of the anticodon arm in translation on the phenotypes of RNA mutants.J Mol Biol. 1986 Nov 20;192(2):235-55. doi: 10.1016/0022-2836(86)90362-1. J Mol Biol. 1986. PMID: 2435916
-
A comprehensive collection of annotations to interpret sequence variation in human mitochondrial transfer RNAs.BMC Bioinformatics. 2016 Nov 8;17(Suppl 12):338. doi: 10.1186/s12859-016-1193-4. BMC Bioinformatics. 2016. PMID: 28185569 Free PMC article.
-
Permutation of a pair of tertiary nucleotides in a transfer RNA.Biochemistry. 1995 Mar 7;34(9):2978-84. doi: 10.1021/bi00009a029. Biochemistry. 1995. PMID: 7534478
-
Pathways to disease from natural variations in human cytoplasmic tRNAs.J Biol Chem. 2019 Apr 5;294(14):5294-5308. doi: 10.1074/jbc.REV118.002982. Epub 2019 Jan 14. J Biol Chem. 2019. PMID: 30643023 Free PMC article. Review.
-
Repertoires of tRNAs: The Couplers of Genomics and Proteomics.Annu Rev Cell Dev Biol. 2018 Oct 6;34:239-264. doi: 10.1146/annurev-cellbio-100617-062754. Epub 2018 Aug 20. Annu Rev Cell Dev Biol. 2018. PMID: 30125138 Review.
Cited by
-
Mistranslating tRNA variants have anticodon- and sex-specific impacts on Drosophila melanogaster.bioRxiv [Preprint]. 2024 Jun 13:2024.06.11.598535. doi: 10.1101/2024.06.11.598535. bioRxiv. 2024. Update in: G3 (Bethesda). 2024 Sep 23:jkae230. doi: 10.1093/g3journal/jkae230. PMID: 38915589 Free PMC article. Updated. Preprint.
-
Noncanonical Roles of tRNAs: tRNA Fragments and Beyond.Annu Rev Genet. 2020 Nov 23;54:47-69. doi: 10.1146/annurev-genet-022620-101840. Epub 2020 Aug 25. Annu Rev Genet. 2020. PMID: 32841070 Free PMC article. Review.
-
Genetic Interaction of tRNA-Dependent Mistranslation with Fused in Sarcoma Protein Aggregates.Genes (Basel). 2023 Feb 18;14(2):518. doi: 10.3390/genes14020518. Genes (Basel). 2023. PMID: 36833445 Free PMC article.
-
A novel mistranslating tRNA model in Drosophila melanogaster has diverse, sexually dimorphic effects.G3 (Bethesda). 2022 May 6;12(5):jkac035. doi: 10.1093/g3journal/jkac035. G3 (Bethesda). 2022. PMID: 35143655 Free PMC article.
-
Impact of tRNA-induced proline-to-serine mistranslation on the transcriptome of Drosophila melanogaster.G3 (Bethesda). 2024 Sep 4;14(9):jkae151. doi: 10.1093/g3journal/jkae151. G3 (Bethesda). 2024. PMID: 38989890 Free PMC article.
References
-
- Hoagland MB, Stephenson ML, Scott JF, et al. A soluble ribonucleic acid intermediate in protein synthesis. J Biol Chem. 1958;231:241–257. - PubMed
-
- Schimmel P. The emerging complexity of the tRNA world: mammalian tRNAs beyond protein synthesis. Nat Rev Mol Cell Biol. 2018;19:45–58. - PubMed
-
- Ling J, Reynolds N, Ibba M. Aminoacyl-tRNA synthesis and translational quality control. Annu Rev Microbiol. 2009;63:61–78. - PubMed
-
- Jenner L, Demeshkina N, Yusupova G, et al. Structural rearrangements of the ribosome at the tRNA proofreading step. Nat Struct Mol Biol. 2010;17:1072–1078. - PubMed
-
- Spahn CMT, Beckmann R, Eswar N, et al. Structure of the 80S ribosome from Saccharomyces cerevisiae - tRNA-ribosome and subunit-subunit interactions. Cell. 2001;107:373–386. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
