A defined human aging phenome
- PMID: 31408848
- PMCID: PMC11627290
- DOI: 10.18632/aging.102166
A defined human aging phenome
Abstract
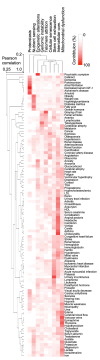
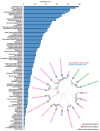
Aging is among the most complex phenotypes that occur in humans. Identifying the interplay between different age-associated features is undoubtedly critical to our understanding of aging and thus age-associated diseases. Nevertheless, what constitutes human aging is not well characterized. Towards this end, we mined millions of PubMed abstracts for age-associated terms, enabling us to generate a detailed description of the human aging phenotype. We discovered age-associated features in clusters that can be broadly associated with previously defined hallmarks of aging, consequently identifying areas where interventions could be pursued. Importantly, we validated the newly discovered features by manually verifying the prevalence of these features in combined cohorts describing 76 million individuals, allowing us to stratify features in aging that appear to be the most prominent. In conclusion, we propose a comprehensive landscape of human aging: the human aging phenome.
Keywords: aging; data mining; phenome; phenotype.
Conflict of interest statement
Figures


References
-
- Mamoshina P, Kochetov K, Putin E, Cortese F, Aliper A, Lee WS, Ahn SM, Uhn L, Skjodt N, Kovalchuk O, Scheibye-Knudsen M, Zhavoronkov A. Population specific biomarkers of human aging: a big data study using South Korean, Canadian and Eastern European patient populations. J Gerontol A Biol Sci Med Sci. 2018; 73:1482–90. 10.1093/gerona/gly005 - DOI - PMC - PubMed
-
- Bobrov E, Georgievskaya A, Kiselev K, Sevastopolsky A, Zhavoronkov A, Gurov S, Rudakov K, Del Pilar Bonilla Tobar M, Jaspers S, Clemann S. PhotoAgeClock: deep learning algorithms for development of non-invasive visual biomarkers of aging. Aging (Albany NY). 2018; 10:3249–59. 10.18632/aging.101629 - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

