A specific class of infectious agents isolated from bovine serum and dairy products and peritumoral colon cancer tissue
- PMID: 31409221
- PMCID: PMC6713099
- DOI: 10.1080/22221751.2019.1651620
A specific class of infectious agents isolated from bovine serum and dairy products and peritumoral colon cancer tissue
Abstract
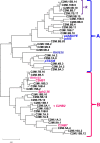
The in silico analyses of 109 replication-competent genomic DNA sequences isolated from cow milk and its products (97 in the bovine meat and milk factors 2 group - BMMF2, and additional 4 in BMMF1) seems to place these in a specific class of infectious agents spanning between bacterial plasmid and circular ssDNA viruses. Satellite-type small plasmids with partial homology to larger genomes, were also isolated in both groups. A member of the BMMF1 group H1MBS.1 was recovered in a distinctly modified form from colon tissue by laser microdissection. Although the evolutionary origin is unknown, it draws the attention to the existence of a hitherto unrecognized, broad spectrum of potential pathogens. Indirect hints to the origin and structure of our isolates, as well as to their replicative behaviour, result from parallels drawn to the Hepatitis deltavirus genome structure and replication.
Keywords: Infectious bovine meat milk factors; colon tissue; new class; pathogenesis; plasmid DNA.
Conflict of interest statement
No potential conflict of interest was reported by the authors.
Figures