GhYGL1d, a pentatricopeptide repeat protein, is required for chloroplast development in cotton
- PMID: 31409298
- PMCID: PMC6693126
- DOI: 10.1186/s12870-019-1945-1
GhYGL1d, a pentatricopeptide repeat protein, is required for chloroplast development in cotton
Abstract
Background: The pentatricopeptide repeat (PPR) gene family, which contains multiple 35-amino acid repeats, constitutes one of the largest gene families in plants. PPR proteins function in organelles to target specific transcripts and are involved in plant development and growth. However, the function of PPR proteins in cotton is still unknown.
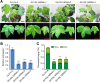
Results: In this study, we characterized a PPR gene YELLOW-GREEN LEAF (GhYGL1d) that is required for cotton plastid development. The GhYGL1d gene has a DYW domain in C-terminal and is highly express in leaves, localized to the chloroplast fractions. GhYGL1d share high amino acid-sequence homology with AtECB2. In atecb2 mutant, overexpression of GhYGL1d rescued the seedling lethal phenotype and restored the editing of accD and ndhF transcripts. Silencing of GhYGL1d led to the reduction of chlorophyll and phenotypically yellow-green leaves in cotton. Compared with wild type, GhYGL1d-silenced cotton showed significant deformations of thylakoid structures. Furthermore, the transcription levels of plastid-encoded polymerase (PEP) and nuclear-encoded polymerase (NEP) dependent genes were decreased in GhYGL1d-silenced cotton.
Conclusions: Our data indicate that GhYGL1d not only contributes to the editing of accD and ndhF genes, but also affects the expression of NEP- and PEP-dependent genes to regulate the development of thylakoids, and therefore regulates leaf variegation in cotton.
Keywords: Chloroplast; Cotton; Leaf variegation; PPR.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures






References
MeSH terms
Substances
Grants and funding
- 31470295/National Natural Science Foundation of China
- 2018JZ3006/Natural Science Basic Research Plan in Shaanxi Province of China
- 2018M640947/Project funded by China Postdoctoral Science Foundation
- GK201901004/Fundamental Research Fund for the Central Universities
- GK201903064/Fundamental Research Fund for the Central Universities
LinkOut - more resources
Full Text Sources
Miscellaneous

