Status and potential of bacterial genomics for public health practice: a scoping review
- PMID: 31409417
- PMCID: PMC6692930
- DOI: 10.1186/s13012-019-0930-2
Status and potential of bacterial genomics for public health practice: a scoping review
Abstract
Background: Next-generation sequencing (NGS) is increasingly being translated into routine public health practice, affecting the surveillance and control of many pathogens. The purpose of this scoping review is to identify and characterize the recent literature concerning the application of bacterial pathogen genomics for public health practice and to assess the added value, challenges, and needs related to its implementation from an epidemiologist's perspective.
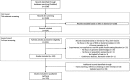
Methods: In this scoping review, a systematic PubMed search with forward and backward snowballing was performed to identify manuscripts in English published between January 2015 and September 2018. Included studies had to describe the application of NGS on bacterial isolates within a public health setting. The studied pathogen, year of publication, country, number of isolates, sampling fraction, setting, public health application, study aim, level of implementation, time orientation of the NGS analyses, and key findings were extracted from each study. Due to a large heterogeneity of settings, applications, pathogens, and study measurements, a descriptive narrative synthesis of the eligible studies was performed.
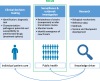
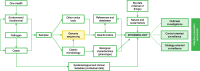
Results: Out of the 275 included articles, 164 were outbreak investigations, 70 focused on strategy-oriented surveillance, and 41 on control-oriented surveillance. Main applications included the use of whole-genome sequencing (WGS) data for (1) source tracing, (2) early outbreak detection, (3) unraveling transmission dynamics, (4) monitoring drug resistance, (5) detecting cross-border transmission events, (6) identifying the emergence of strains with enhanced virulence or zoonotic potential, and (7) assessing the impact of prevention and control programs. The superior resolution over conventional typing methods to infer transmission routes was reported as an added value, as well as the ability to simultaneously characterize the resistome and virulome of the studied pathogen. However, the full potential of pathogen genomics can only be reached through its integration with high-quality contextual data.
Conclusions: For several pathogens, it is time for a shift from proof-of-concept studies to routine use of WGS during outbreak investigations and surveillance activities. However, some implementation challenges from the epidemiologist's perspective remain, such as data integration, quality of contextual data, sampling strategies, and meaningful interpretations. Interdisciplinary, inter-sectoral, and international collaborations are key for an appropriate genomics-informed surveillance.
Keywords: Bacterial infections; Epidemiology; Genomics; Next-generation sequencing; Public health practice; Scoping review; Whole-genome sequencing.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
References
-
- Struelens MJ, Brisse S. From molecular to genomic epidemiology: transforming surveillance and control of infectious diseases. Eurosurveillance. 2013;18(4):pii = 20386. Available from: http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId = 20386 - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

