Engineered Interspecies Amino Acid Cross-Feeding Increases Population Evenness in a Synthetic Bacterial Consortium
- PMID: 31409662
- PMCID: PMC6697442
- DOI: 10.1128/mSystems.00352-19
Engineered Interspecies Amino Acid Cross-Feeding Increases Population Evenness in a Synthetic Bacterial Consortium
Abstract
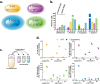
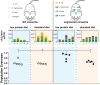
In nature, microbes interact antagonistically, neutrally, or beneficially. To shed light on the effects of positive interactions in microbial consortia, we introduced metabolic dependencies and metabolite overproduction into four bacterial species. While antagonistic interactions govern the wild-type consortium behavior, the genetic modifications alleviated antagonistic interactions and resulted in beneficial interactions. Engineered cross-feeding increased population evenness, a component of ecological diversity, in different environments, including in a more complex gnotobiotic mouse gut environment. Our findings suggest that metabolite cross-feeding could be used as a tool for intentionally shaping microbial consortia in complex environments.IMPORTANCE Microbial communities are ubiquitous in nature. Bacterial consortia live in and on our body and in our environment, and more recently, biotechnology is applying microbial consortia for bioproduction. As part of our body, bacterial consortia influence us in health and disease. Microbial consortium function is determined by its composition, which in turn is driven by the interactions between species. Further understanding of microbial interactions will help us in deciphering how consortia function in complex environments and may enable us to modify microbial consortia for health and environmental benefits.
Keywords: metabolite cross-feeding; microbial consortia; synthetic biology.
Copyright © 2019 Ziesack et al.
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

