The Cervicovaginal Microbiota-Host Interaction Modulates Chlamydia trachomatis Infection
- PMID: 31409678
- PMCID: PMC6692509
- DOI: 10.1128/mBio.01548-19
The Cervicovaginal Microbiota-Host Interaction Modulates Chlamydia trachomatis Infection
Abstract
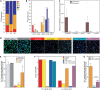
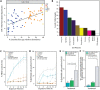
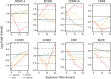
The mechanism(s) by which Lactobacillus-dominated cervicovaginal microbiota provide a barrier to Chlamydia trachomatis infection remain(s) unknown. Here we evaluate the impact of different Lactobacillus spp. identified via culture-independent metataxonomic analysis of C. trachomatis-infected women on C. trachomatis infection in a three-dimensional (3D) cervical epithelium model. Lactobacillus spp. that specifically produce d(-) lactic acid were associated with long-term protection against C. trachomatis infection, consistent with reduced protection associated with Lactobacillus iners, which does not produce this isoform, and with decreased epithelial cell proliferation, consistent with the observed prolonged protective effect. Transcriptomic analysis revealed that epigenetic modifications involving histone deacetylase-controlled pathways are integral to the cross talk between host and microbiota. These results highlight a fundamental mechanism whereby the cervicovaginal microbiota modulates host functions to protect against C. trachomatis infection.IMPORTANCE The vaginal microbiota is believed to protect women against Chlamydia trachomatis, the etiologic agent of the most prevalent sexually transmitted infection (STI) in developed countries. The mechanism underlying this protection has remained elusive. Here, we reveal the comprehensive strategy by which the cervicovaginal microbiota modulates host functions to protect against chlamydial infection, thereby providing a novel conceptual mechanistic understanding. Major implications of this work are that (i) the impact of the vaginal microbiota on the epithelium should be considered in future studies of chlamydial infection and other STIs and (ii) a fundamental understanding of the cervicovaginal microbiota's role in protection against STIs may enable the development of novel microbiome-based therapeutic strategies to protect women from infection and improve vaginal and cervical health.
Keywords: Lactobacillus; epigenetic; lactic acid; microbiome; proliferation; sexually transmitted infection.
Copyright © 2019 Edwards et al.
Figures


References
-
- Newman L, Rowley J, Vander Hoorn S, Wijesooriya NS, Unemo M, Low N, Stevens G, Gottlieb S, Kiarie J, Temmerman M. 2015. Global estimates of the prevalence and incidence of four curable sexually transmitted infections in 2012 based on systematic review and global reporting. PLoS One 10:e0143304. doi:10.1371/journal.pone.0143304. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous
