A chromosome-anchored eggplant genome sequence reveals key events in Solanaceae evolution
- PMID: 31409808
- PMCID: PMC6692341
- DOI: 10.1038/s41598-019-47985-w
A chromosome-anchored eggplant genome sequence reveals key events in Solanaceae evolution
Abstract
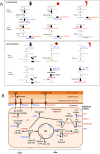
With approximately 450 species, spiny Solanum species constitute the largest monophyletic group in the Solanaceae family, but a high-quality genome assembly from this group is presently missing. We obtained a chromosome-anchored genome assembly of eggplant (Solanum melongena), containing 34,916 genes, confirming that the diploid gene number in the Solanaceae is around 35,000. Comparative genomic studies with tomato (S. lycopersicum), potato (S. tuberosum) and pepper (Capsicum annuum) highlighted the rapid evolution of miRNA:mRNA regulatory pairs and R-type defense genes in the Solanaceae, and provided a genomic basis for the lack of steroidal glycoalkaloid compounds in the Capsicum genus. Using parsimony methods, we reconstructed the putative chromosomal complements of the key founders of the main Solanaceae clades and the rearrangements that led to the karyotypes of extant species and their ancestors. From 10% to 15% of the genes present in the four genomes were syntenic paralogs (ohnologs) generated by the pre-γ, γ and T paleopolyploidy events, and were enriched in transcription factors. Our data suggest that the basic gene network controlling fruit ripening is conserved in different Solanaceae clades, and that climacteric fruit ripening involves a differential regulation of relatively few components of this network, including CNR and ethylene biosynthetic genes.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Vavilov N. The origin, variation, immunity and breeding of cultivated plants. English Transl. by K.S. Chester. Chron. Bot. 1951;13:1–366.

