Genome-wide association analysis of self-reported daytime sleepiness identifies 42 loci that suggest biological subtypes
- PMID: 31409809
- PMCID: PMC6692391
- DOI: 10.1038/s41467-019-11456-7
Genome-wide association analysis of self-reported daytime sleepiness identifies 42 loci that suggest biological subtypes
Abstract
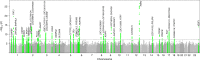
Excessive daytime sleepiness (EDS) affects 10-20% of the population and is associated with substantial functional deficits. Here, we identify 42 loci for self-reported daytime sleepiness in GWAS of 452,071 individuals from the UK Biobank, with enrichment for genes expressed in brain tissues and in neuronal transmission pathways. We confirm the aggregate effect of a genetic risk score of 42 SNPs on daytime sleepiness in independent Scandinavian cohorts and on other sleep disorders (restless legs syndrome, insomnia) and sleep traits (duration, chronotype, accelerometer-derived sleep efficiency and daytime naps or inactivity). However, individual daytime sleepiness signals vary in their associations with objective short vs long sleep, and with markers of sleep continuity. The 42 sleepiness variants primarily cluster into two predominant composite biological subtypes - sleep propensity and sleep fragmentation. Shared genetic links are also seen with obesity, coronary heart disease, psychiatric diseases, cognitive traits and reproductive ageing.
Conflict of interest statement
H.O. is a consultant for Jazz Pharmaceuticals, Medix Biochemica, and Roche Holding. K. Kristiansson is a Consultant for Negen Ltd. F.A.J.L.S. has reveived speaker fees from Bayer Healthcare, Sentara Healthcare, Philips, Kellogg Company, Vanda Pharmaceuticals, and Pfizer. M.K.R. has acted as a consultant for Novo Nordisk and Roche Diabetes Care, and also participated in advisory board meetings on their behalf. The remaining authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Grants and funding
- MR/P023576/2/MRC_/Medical Research Council/United Kingdom
- MC_UU_00011/2/MRC_/Medical Research Council/United Kingdom
- MC_QA137853/MRC_/Medical Research Council/United Kingdom
- MR/P012167/1/MRC_/Medical Research Council/United Kingdom
- R01 HL113338/HL/NHLBI NIH HHS/United States
- R01 HL127564/HL/NHLBI NIH HHS/United States
- R35 HL135818/HL/NHLBI NIH HHS/United States
- 107851/Z/15/Z/WT_/Wellcome Trust/United Kingdom
- F32 DK102323/DK/NIDDK NIH HHS/United States
- MR/M008908/1/MRC_/Medical Research Council/United Kingdom
- MC_UU_00011/3/MRC_/Medical Research Council/United Kingdom
- R01 HG003054/HG/NHGRI NIH HHS/United States
- MR/P023576/1/MRC_/Medical Research Council/United Kingdom
- R35 HL135824/HL/NHLBI NIH HHS/United States
- MR/M005070/1/MRC_/Medical Research Council/United Kingdom
- K01 HL136884/HL/NHLBI NIH HHS/United States
- R01 DK107859/DK/NIDDK NIH HHS/United States
- R01 DK102696/DK/NIDDK NIH HHS/United States
- MC_UU_00011/6/MRC_/Medical Research Council/United Kingdom
- MC_PC_17228/MRC_/Medical Research Council/United Kingdom
- T32 HL007567/HL/NHLBI NIH HHS/United States
- K01 HL135405/HL/NHLBI NIH HHS/United States
- R01 DK105072/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

