Comparative Proteomic Analysis Provides Insights into the Regulatory Mechanisms of Wheat Primary Root Growth
- PMID: 31409818
- PMCID: PMC6692329
- DOI: 10.1038/s41598-019-47926-7
Comparative Proteomic Analysis Provides Insights into the Regulatory Mechanisms of Wheat Primary Root Growth
Abstract
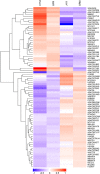
Plant roots are vital for acquiring nutrients and water from soil. However, the mechanisms regulating root growth in hexaploid wheat remain to be elucidated. Here, an integrated comparative proteome study on the roots of two varieties and their descendants with contrasting root phenotypes was performed. A total of 80 differentially expressed proteins (DEPs) associated with the regulation of primary root growth were identified, including two plant steroid biosynthesis related proteins and nine class III peroxidases. Real-time PCR analysis showed that brassinosteroid (BR) biosynthesis pathway was significantly elevated in long-root plants compared with those short-root plants. Moreover, O2.- and H2O2 were distributed abundantly in both the root meristematic and elongation zones of long root plants, but only in the meristematic zone of short-root plants. The differential distribution of reactive oxygen species (ROS) in the root tips of different genotypes may be caused by the differential expression of peroxidases. Taken together, our results suggest that the regulation of wheat primary root growth is closely related to BR biosynthesis pathway and BR-mediated ROS distribution.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Rosegrant MW, Ringler C, Zhu T. Water for agriculture: maintaining food security under growing scarcity. Annu. Rev. Environ. Resour. 2009;34:205–222. doi: 10.1146/annurev.environ.030308.090351. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

