Centrifugation-Assisted Immiscible Fluid Filtration for Dual-Bioanalyte Extraction
- PMID: 31411020
- PMCID: PMC7521759
- DOI: 10.1021/acs.analchem.9b02572
Centrifugation-Assisted Immiscible Fluid Filtration for Dual-Bioanalyte Extraction
Abstract
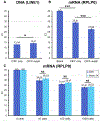
The extraction of bioanalytes is the first step in many diagnostic and analytical assays. However, most bioanalyte extraction methods require extensive dilution-based washing processes that are not only time-consuming and laborious but can also result in significant sample loss, limiting their applications in rare sample analyses. Here, we present a method that enables the efficient extraction of multiple different bioanalytes from rare samples (down to 10 cells) without washing-centrifugation-assisted immiscible fluid filtration (CIFF). CIFF utilizes centrifugal force to drive the movement of analyte-bound glass microbeads from an aqueous sample into an immiscible hydrophobic solution to perform an efficient, simple, and nondilutive extraction. The method can be performed using conventional polymerase chain reaction (PCR) tubes with no requirement of specialized devices, columns, or instruments, making it broadly accessible and cost-effective. The CIFF process can effectively remove approximately 99.5% of the aqueous sample in one extraction with only 0.5% residual carryover, whereas a traditional "spin-down and aspirate" operation results in a higher 3.6% carryover. Another unique aspect of CIFF is its ability to perform two different solid-phase bioanalytes extractions simultaneously within a single vessel without fractionating the sample or performing serial extractions. Here we demonstrate efficient mRNA and DNA extraction from low-input samples (down to 10 cells) with slightly higher to comparable recovery compared to a traditional column-based extraction technique and the simultaneous extraction of two different proteins in the same tube using CIFF.
Figures
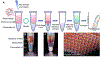


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

