PIEZO1 activation delays erythroid differentiation of normal and hereditary xerocytosis-derived human progenitor cells
- PMID: 31413092
- PMCID: PMC7049340
- DOI: 10.3324/haematol.2019.218503
PIEZO1 activation delays erythroid differentiation of normal and hereditary xerocytosis-derived human progenitor cells
Abstract
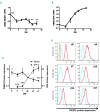
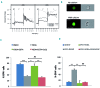
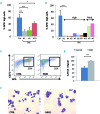
Hereditary xerocytosis is a dominantly inherited red cell membrane disorder caused in most cases by gain-of-function mutations in PIEZO1, encoding a mechanosensitive ion channel that translates a mechanic stimulus into calcium influx. We found that PIEZO1 was expressed early in erythroid progenitor cells, and investigated whether it could be involved in erythropoiesis, besides having a role in the homeostasis of mature red cell hydration. In UT7 cells, chemical PIEZO1 activation using YODA1 repressed glycophorin A expression by 75%. This effect was PIEZO1-dependent since it was reverted using specific short hairpin-RNA knockdown. The effect of PIEZO1 activation was confirmed in human primary progenitor cells, maintaining cells at an immature stage for longer and modifying the transcriptional balance in favor of genes associated with early erythropoiesis, as shown by a high GATA2/GATA1 ratio and decreased α/β-globin expression. The cell proliferation rate was also reduced, with accumulation of cells in G0/G1 of the cell cycle. The PIEZO1-mediated effect on UT7 cells required calcium-dependent activation of the NFAT and ERK1/2 pathways. In primary erythroid cells, PIEZO1 activation synergized with erythropoietin to activate STAT5 and ERK, indicating that it may modulate signaling pathways downstream of erythropoietin receptor activation. Finally, we studied the in-vitro erythroid differentiation of primary cells obtained from 14 PIEZO1-mutated patients, from 11 families, carrying ten different mutations. We observed a delay in erythroid differentiation in all cases, ranging from mild (n=3) to marked (n=8). Overall, these data demonstrate a role for PIEZO1 during erythropoiesis, since activation of PIEZO1 - both chemically and through activating mutations - delays erythroid maturation, providing new insights into the pathophysiology of hereditary xerocytosis.
Copyright© 2020 Ferrata Storti Foundation.
Figures




Comment in
-
Staying hydrated is important also for erythroblasts.Haematologica. 2020 Mar;105(3):528-529. doi: 10.3324/haematol.2019.233999. Haematologica. 2020. PMID: 32115411 Free PMC article. No abstract available.
References
-
- Zhao Q, Zhou H, Chi S, et al. Structure and mechanogating mechanism of the Piezo1 channel.Nature.2018;554(7693):487–492. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Miscellaneous

