Phylogeography of Lassa Virus in Nigeria
- PMID: 31413134
- PMCID: PMC6803284
- DOI: 10.1128/JVI.00929-19
Phylogeography of Lassa Virus in Nigeria
Abstract
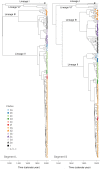
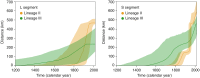
Lassa virus is genetically diverse with several lineages circulating in West Africa. This study aimed at describing the sequence variability of Lassa virus across Nigeria and inferring its spatiotemporal evolution. We sequenced and isolated 77 Lassa virus strains from 16 Nigerian states. The final data set, including previous works, comprised metadata and sequences of 219 unique strains sampled between 1969 and 2018 in 22 states. Most of this data originated from Lassa fever patients diagnosed at Irrua Specialist Teaching Hospital, Edo State, Nigeria. The majority of sequences clustered with the main Nigerian lineages II and III, while a few sequences formed a new cluster related to Lassa virus strains from Hylomyscus pamfi Within lineages II and III, seven and five sublineages, respectively, were distinguishable. Phylogeographic analysis suggests an origin of lineage II in the southeastern part of the country around Ebonyi State and a main vector of dispersal toward the west across the Niger River, through Anambra, Kogi, Delta, and Edo into Ondo State. The frontline of virus dispersal appears to be in Ondo. Minor vectors are directed northeast toward Taraba and Adamawa and south toward Imo and Rivers. Lineage III might have spread from northern Plateau State into Kaduna, Nasarawa, Federal Capital Territory, and Bauchi. One sublineage moved south and crossed the Benue River into Benue State. This study provides a geographic mapping of lineages and phylogenetic clusters in Nigeria at a higher resolution. In addition, we estimated the direction and time frame of virus dispersal in the country.IMPORTANCE Lassa virus is the causative agent of Lassa fever, a viral hemorrhagic fever with a case fatality rate of approximately 30% in Africa. Previous studies disclosed a geographical pattern in the distribution of Lassa virus strains and a westward movement of the virus across West Africa during evolution. Our study provides a deeper understanding of the geography of genetic lineages and sublineages of the virus in Nigeria. In addition, we modeled how the virus spread in the country. This knowledge allows us to predict into which geographical areas the virus might spread in the future and prioritize areas for Lassa fever surveillance. Our study not only aimed to generate Lassa virus sequences from across Nigeria but also to isolate and conserve the respective viruses for future research. Both isolates and sequences are important for the development and evaluation of medical countermeasures to treat and prevent Lassa fever, such as diagnostics, therapeutics, and vaccines.
Keywords: Lassa virus; Nigeria; phylogeny.
Copyright © 2019 Ehichioya et al.
Figures

References
-
- Okokhere P, Colubri A, Azubike C, Iruolagbe C, Osazuwa O, Tabrizi S, Chin E, Asad S, Ediale E, Rafiu M, Adomeh D, Odia I, Atafo R, Aire C, Okogbenin S, Pahlman M, Becker-Ziaja B, Asogun D, Fradet T, Fry B, Schaffner SF, Happi C, Akpede G, Günther S, Sabeti PC. 2018. Clinical and laboratory predictors of Lassa fever outcome in a dedicated treatment facility in Nigeria: a retrospective, observational cohort study. Lancet Infect Dis 18:684–695. doi:10.1016/S1473-3099(18)30121-X. - DOI - PMC - PubMed
-
- Asogun DA, Adomeh DI, Ehimuan J, Odia I, Hass M, Gabriel M, Olschläger S, Becker-Ziaja B, Folarin O, Phelan E, Ehiane PE, Ifeh VE, Uyigue EA, Oladapo YT, Muoebonam EB, Osunde O, Dongo A, Okokhere PO, Okogbenin SA, Momoh M, Alikah SO, Akhuemokhan OC, Imomeh P, Odike MAC, Gire S, Andersen K, Sabeti PC, Happi CT, Akpede GO, Günther S. 2012. Molecular diagnostics for Lassa fever at Irrua specialist teaching hospital, Nigeria: lessons learnt from two years of laboratory operation. PLoS Negl Trop Dis 6:e1839. doi:10.1371/journal.pntd.0001839. - DOI - PMC - PubMed
-
- Shaffer JG, Grant DS, Schieffelin JS, Boisen ML, Goba A, Hartnett JN, Levy DC, Yenni RE, Moses LM, Fullah M, Momoh M, Fonnie M, Fonnie R, Kanneh L, Koroma VJ, Kargbo K, Ottomassathien D, Muncy IJ, Jones AB, Illick MM, Kulakosky PC, Haislip AM, Bishop CM, Elliot DH, Brown BL, Zhu H, Hastie KM, Andersen KG, Gire SK, Tabrizi S, Tariyal R, Stremlau M, Matschiner A, Sampey DB, Spence JS, Cross RW, Geisbert JB, Folarin OA, Happi CT, Pitts KR, Geske FJ, Geisbert TW, Saphire EO, Robinson JE, Wilson RB, Sabeti PC, Henderson LA, Khan SH, Bausch DG, Branco LM, Garry RF, Viral Hemorrhagic Fever Consortium. 2014. Lassa fever in post-conflict Sierra Leone. PLoS Negl Trop Dis 8:e2748. doi:10.1371/journal.pntd.0002748. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

