Hydrophobic catalysis and a potential biological role of DNA unstacking induced by environment effects
- PMID: 31413203
- PMCID: PMC6717297
- DOI: 10.1073/pnas.1909122116
Hydrophobic catalysis and a potential biological role of DNA unstacking induced by environment effects
Abstract
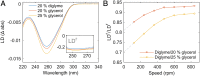
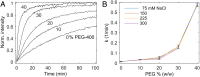
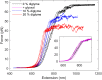
Hydrophobic base stacking is a major contributor to DNA double-helix stability. We report the discovery of specific unstacking effects in certain semihydrophobic environments. Water-miscible ethylene glycol ethers are found to modify structure, dynamics, and reactivity of DNA by mechanisms possibly related to a biologically relevant hydrophobic catalysis. Spectroscopic data and optical tweezers experiments show that base-stacking energies are reduced while base-pair hydrogen bonds are strengthened. We propose that a modulated chemical potential of water can promote "longitudinal breathing" and the formation of unstacked holes while base unpairing is suppressed. Flow linear dichroism in 20% diglyme indicates a 20 to 30% decrease in persistence length of DNA, supported by an increased flexibility in single-molecule nanochannel experiments in poly(ethylene glycol). A limited (3 to 6%) hyperchromicity but unaffected circular dichroism is consistent with transient unstacking events while maintaining an overall average B-DNA conformation. Further information about unstacking dynamics is obtained from the binding kinetics of large thread-intercalating ruthenium complexes, indicating that the hydrophobic effect provides a 10 to 100 times increased DNA unstacking frequency and an "open hole" population on the order of 10-2 compared to 10-4 in normal aqueous solution. Spontaneous DNA strand exchange catalyzed by poly(ethylene glycol) makes us propose that hydrophobic residues in the L2 loop of recombination enzymes RecA and Rad51 may assist gene recombination via modulation of water activity near the DNA helix by hydrophobic interactions, in the manner described here. We speculate that such hydrophobic interactions may have catalytic roles also in other biological contexts, such as in polymerases.
Keywords: DNA; DNA polymerase; RecA; hydrophobic catalysis; threading intercalation.
Copyright © 2019 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Mak C. H., Unraveling base stacking driving forces in DNA. J. Phys. Chem. B 120, 6010–6020 (2016). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

