The PINK1 kinase-driven ubiquitin ligase Parkin promotes mitochondrial protein import through the presequence pathway in living cells
- PMID: 31413265
- PMCID: PMC6694185
- DOI: 10.1038/s41598-019-47352-9
The PINK1 kinase-driven ubiquitin ligase Parkin promotes mitochondrial protein import through the presequence pathway in living cells
Abstract
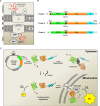
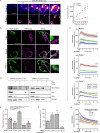
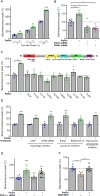
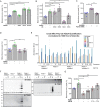
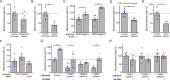
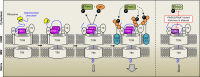
Most of over a thousand mitochondrial proteins are encoded by nuclear genes and must be imported from the cytosol. Little is known about the cytosolic events regulating mitochondrial protein import, partly due to the lack of appropriate tools for its assessment in living cells. We engineered an inducible biosensor for monitoring the main presequence-mediated import pathway with a quantitative, luminescence-based readout. This tool was used to explore the regulation of mitochondrial import by the PINK1 kinase-driven Parkin ubiquitin ligase, which is dysfunctional in autosomal recessive Parkinson's disease. We show that mitochondrial import was stimulated by Parkin, but not by disease-causing Parkin variants. This effect was dependent on Parkin activation by PINK1 and accompanied by an increase in the abundance of K11 ubiquitin chains on mitochondria and by ubiquitylation of subunits of the translocase of outer mitochondrial membrane. Mitochondrial import efficiency was abnormally low in cells from patients with PINK1- and PARK2-linked Parkinson's disease and was restored by phosphomimetic ubiquitin in cells with residual Parkin activity. Altogether, these findings uncover a role of ubiquitylation in mitochondrial import regulation and suggest that loss of this regulatory loop may underlie the pathophysiology of Parkinson's disease, providing novel opportunities for therapeutic intervention.
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
The TOMM machinery is a molecular switch in PINK1 and PARK2/PARKIN-dependent mitochondrial clearance.Autophagy. 2013 Nov 1;9(11):1801-17. doi: 10.4161/auto.25884. Epub 2013 Sep 5. Autophagy. 2013. PMID: 24149440
-
PINK1 import regulation at a crossroad of mitochondrial fate: the molecular mechanisms of PINK1 import.J Biochem. 2020 Mar 1;167(3):217-224. doi: 10.1093/jb/mvz069. J Biochem. 2020. PMID: 31504668 Review.
-
Parkin maintains mitochondrial levels of the protective Parkinson's disease-related enzyme 17-β hydroxysteroid dehydrogenase type 10.Cell Death Differ. 2015 Oct;22(10):1563-76. doi: 10.1038/cdd.2014.224. Epub 2015 Jan 16. Cell Death Differ. 2015. PMID: 25591737 Free PMC article.
-
Defining roles of PARKIN and ubiquitin phosphorylation by PINK1 in mitochondrial quality control using a ubiquitin replacement strategy.Proc Natl Acad Sci U S A. 2015 May 26;112(21):6637-42. doi: 10.1073/pnas.1506593112. Epub 2015 May 12. Proc Natl Acad Sci U S A. 2015. PMID: 25969509 Free PMC article.
-
Mechanisms of mitophagy: PINK1, Parkin, USP30 and beyond.Free Radic Biol Med. 2016 Nov;100:210-222. doi: 10.1016/j.freeradbiomed.2016.04.015. Epub 2016 Apr 16. Free Radic Biol Med. 2016. PMID: 27094585 Review.
Cited by
-
PINK1/Parkin Mediated Mitophagy, Ca2+ Signalling, and ER-Mitochondria Contacts in Parkinson's Disease.Int J Mol Sci. 2020 Mar 5;21(5):1772. doi: 10.3390/ijms21051772. Int J Mol Sci. 2020. PMID: 32150829 Free PMC article. Review.
-
The Mitochondrial Kinase PINK1 in Diabetic Kidney Disease.Int J Mol Sci. 2021 Feb 3;22(4):1525. doi: 10.3390/ijms22041525. Int J Mol Sci. 2021. PMID: 33546409 Free PMC article. Review.
-
A Biochemical and Structural Understanding of TOM Complex Interactions and Implications for Human Health and Disease.Cells. 2021 May 11;10(5):1164. doi: 10.3390/cells10051164. Cells. 2021. PMID: 34064787 Free PMC article. Review.
-
Mitochondria in neurodegeneration.Curr Opin Physiol. 2022 Apr;26:100532. doi: 10.1016/j.cophys.2022.100532. Epub 2022 Apr 1. Curr Opin Physiol. 2022. PMID: 35814636 Free PMC article.
-
Mitochondrial quality control: from molecule to organelle.Cell Mol Life Sci. 2021 Apr;78(8):3853-3866. doi: 10.1007/s00018-021-03775-0. Epub 2021 Mar 29. Cell Mol Life Sci. 2021. PMID: 33782711 Free PMC article. Review.
References
-
- Devi L, Prabhu BM, Galati DF, Avadhani NG, Anandatheerthavarada HK. Accumulation of amyloid precursor protein in the mitochondrial import channels of human Alzheimer’s disease brain is associated with mitochondrial dysfunction. J Neurosci. 2006;26:9057–9068. doi: 10.1523/JNEUROSCI.1469-06.2006. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

