Advances in gene ontology utilization improve statistical power of annotation enrichment
- PMID: 31415589
- PMCID: PMC6695228
- DOI: 10.1371/journal.pone.0220728
Advances in gene ontology utilization improve statistical power of annotation enrichment
Abstract
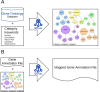
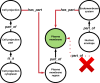
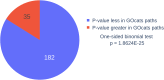
Gene-annotation enrichment is a common method for utilizing ontology-based annotations in gene and gene-product centric knowledgebases. Effective utilization of these annotations requires inferring semantic linkages by tracing paths through edges in the ontological graph, referred to as relations. However, some relations are semantically problematic with respect to scope, necessitating their omission or modification lest erroneous term mappings occur. To address these issues, we created the Gene Ontology Categorization Suite, or GOcats-a novel tool that organizes the Gene Ontology into subgraphs representing user-defined concepts, while ensuring that all appropriate relations are congruent with respect to scoping semantics. Here, we demonstrate the improvements in annotation enrichment by re-interpreting edges that would otherwise be omitted by traditional ancestor path-tracing methods. Specifically, we show that GOcats' unique handling of relations improves enrichment over conventional methods in the analysis of two different gene-expression datasets: a breast cancer microarray dataset and several horse cartilage development RNAseq datasets. With the breast cancer microarray dataset, we observed significant improvement (one-sided binomial test p-value = 1.86E-25) in 182 of 217 significantly enriched GO terms identified from the conventional path traversal method when GOcats' path traversal was used. We also found new significantly enriched terms using GOcats, whose biological relevancy has been experimentally demonstrated elsewhere. Likewise, on the horse RNAseq datasets, we observed a significant improvement in GO term enrichment when using GOcat's path traversal: one-sided binomial test p-values range from 1.32E-03 to 2.58E-44.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

