Phylogenetic analysis revealed the co-circulation of four dengue virus serotypes in Southern Thailand
- PMID: 31415663
- PMCID: PMC6695175
- DOI: 10.1371/journal.pone.0221179
Phylogenetic analysis revealed the co-circulation of four dengue virus serotypes in Southern Thailand
Abstract
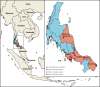
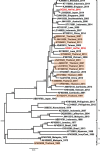
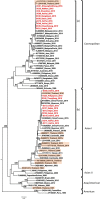
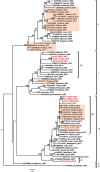
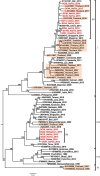
Dengue fever is caused by dengue viruses (DENV) from the Flavivirus genus and is the most prevalent arboviral disease. DENV exists in four immunogenically distinct and genetically-related serotypes (DENV-1 to 4), each subdivided in genotypes. Despite the endemicity of all four DENV serotypes in Thailand, no prior study has characterized the circulation of DENV in the southern provinces of the country. To determine the genetic diversity of DENV circulating in Southern Thailand in 2015 and 2016, we investigated 46 viruses from 182 patients' sera confirmed positive for DENV by serological and Nested RT-PCR tests. Our dataset included 2 DENV-1, 20 DENV-2, 9 DENV-3 and 15 DENV-4. Phylogenetic analysis was performed on viral envelop sequences. This revealed that part of the identified genotypes from DENV-1 and DENV-4 had been predominant in Asia (genotype I for both serotypes), while genotype II for DENV-4 and the Cosmopolitan genotype DENV-2 were also circulating. Whereas DENV-3 genotype II had been predominantly detected in South East Asia during the previous decades, we found genotype III and genotype I in Southern Thailand. All DENV genotype identified in this study were closely related to contemporary strains circulating in Southeast Asian countries, emphasizing the regional circulation of DENV. These results provide new insights into the co-circulation of all four DENV serotypes in Southern Thailand, confirming the hyperendemicity of DENV in the region. These findings also suggest a new trend of dissemination for some DENV serotypes with a possible shift in genotype distribution; as recently observed in other Asian countries.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
References
-
- WHO. Dengue and severe dengue 2018. Available from: http://www.who.int/news-room/fact-sheets/detail/dengue-and-severe-dengue.
-
- Flasche S, Jit M, Rodríguez-Barraquer I, Coudeville L, Recker M, Koelle K, et al. The Long-Term Safety, Public Health Impact, and Cost-Effectiveness of Routine Vaccination with a Recombinant, Live-Attenuated Dengue Vaccine (Dengvaxia): A Model Comparison Study. PLoS Med. 2016;13(11):e1002181 Epub 2016/11/29. 10.1371/journal.pmed.1002181 - DOI - PMC - PubMed
-
- Holmes EC, Burch SS. The causes and consequences of genetic variation in dengue virus. Trends Microbiol. 2000;8(2):74–7. . - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

