Universal principles of membrane protein assembly, composition and evolution
- PMID: 31415673
- PMCID: PMC6695178
- DOI: 10.1371/journal.pone.0221372
Universal principles of membrane protein assembly, composition and evolution
Abstract
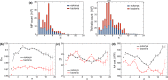
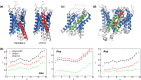
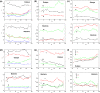
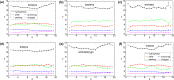
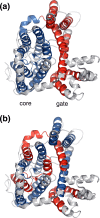
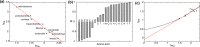
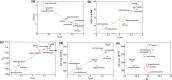
Structural diversity in α-helical membrane proteins (MP) arises from variations in helix-helix crossings and contacts that may bias amino acid usage. Here, we reveal systematic changes in transmembrane amino acid frequencies (f) as a function of the number of helices (n). For eukarya, breaks in f(n) trends of packing (Ala, Gly and Pro), polar, and hydrophobic residues identify different MP assembly principles for 2≤n≤7, 8≤n≤12 and n≥13. In bacteria, the first f break already occurs after n = 6 in correlation to an earlier n peak in MP size distribution and dominance of packing over polar interactions. In contrast to the later n brackets, the integration levels of helix bundles continuously increased in the first, most populous brackets indicating the formation of single structural units (domains). The larger first bracket of eukarya relates to a balance of polar and packing interactions that enlarges helix-helix combinatorial possibilities (MP diversity). Between the evolutionary old, packing and new, polar residues f anti-correlations extend over all biological taxa, broadly ordering them according to evolutionary history and allowing f estimates for the earliest forms of life. Next to evolutionary history, the amino acid composition of MP is determined by size (n), proteome diversity, and effective amino acid cost.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

