Bacterial-Fungal Interactions in the Kelp Endomicrobiota Drive Autoinducer-2 Quorum Sensing
- PMID: 31417510
- PMCID: PMC6685064
- DOI: 10.3389/fmicb.2019.01693
Bacterial-Fungal Interactions in the Kelp Endomicrobiota Drive Autoinducer-2 Quorum Sensing
Abstract
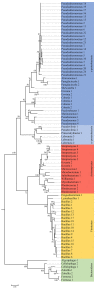
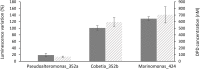
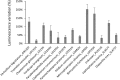
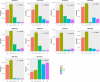
Brown macroalgae are an essential component of temperate coastal ecosystems and a growing economic sector. They harbor diverse microbial communities that regulate algal development and health. This algal holobiont is dynamic and achieves equilibrium via a complex network of microbial and host interactions. We now report that bacterial and fungal endophytes associated with four brown algae (Ascophyllum nodosum, Pelvetia canaliculata, Laminaria digitata, and Saccharina latissima) produce metabolites that interfere with bacterial autoinducer-2 quorum sensing, a signaling system implicated in virulence and host colonization. Additionally, we performed co-culture experiments combined to a metabolomic approach and demonstrated that microbial interactions influence production of metabolites, including metabolites involved in quorum sensing. Collectively, the data highlight autoinducer-2 quorum sensing as a key metabolite in the complex network of interactions within the algal holobiont.
Keywords: AI-2; algal holobiont; bacterial–fungal interaction; kelp microbiota; quorum sensing (QS).
Figures






References
-
- Albakosh M. A., Naidoo R. K., Kirby M., Bauer R. (2016). Identification of epiphytic bacterial communities associated with the brown alga Splachnidium rugosum. J. Appl. Phycol. 28 1891–1901. 10.1007/s10811-015-0725-z - DOI
LinkOut - more resources
Full Text Sources

