In-Frame Indel Mutations in the Genome of the Blind Mexican Cavefish, Astyanax mexicanus
- PMID: 31418011
- PMCID: PMC6751357
- DOI: 10.1093/gbe/evz180
In-Frame Indel Mutations in the Genome of the Blind Mexican Cavefish, Astyanax mexicanus
Abstract
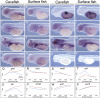
Organisms living in the subterranean biome evolve extreme characteristics including vision loss and sensory expansion. Despite prior work linking certain genes to Mendelian traits, the genetic basis for complex cave-associated traits remains unknown. Moreover, it is unclear if certain forms of genetic variation (e.g., indels, copy number variants) are more common in regressive evolution. Progress in this area has been limited by a lack of suitable natural model systems and genomic resources. In recent years, the Mexican tetra, Astyanax mexicanus, has advanced as a model for cave biology and regressive evolution. Here, we present the results of a genome-wide screen for in-frame indels using alignments of RNA-sequencing reads to the draft cavefish genome. Mutations were discovered in three genes associated with blood physiology (mlf1, plg, and wdr1), two genes associated with growth factor signaling (ghrb, rnf126), one gene linked to collagen defects (mia3), and one gene which may have a global epigenetic impact on gene expression (mki67). With one exception, polymorphisms were shared between Pachón and Tinaja cavefish lineages, and different from the surface-dwelling lineage. We confirmed the presence of mutations using direct Sanger sequencing and discovered remarkably similar developmental expression in both morphs despite substantial coding sequence alterations. Further, three mutated genes mapped near previously established quantitative trait loci associated with jaw size, condition factor, lens size, and neuromast variation. This work reveals previously unappreciated traits evolving in this species under environmental pressures (e.g., blood physiology) and provides insight to genetic changes underlying convergence of organisms evolving in complete darkness.
Keywords: Astyanax; RNA-sequencing; genetic lesion; troglomorphy.
© The Author(s) 2019. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures


References
-
- Dowling TE, Martasian DP, Jeffery WR.. 2002. Evidence for multiple genetic forms with similar eyeless phenotypes in the blind cavefish, Astyanax mexicanus. Mol Biol Evol. 19(4):446–455. - PubMed
-
- Drew AF, et al. 1998. Ligneous conjunctivitis in plasminogen-deficient mice. Blood 91(5):1616–1624. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

