Diversification of Retinoblastoma Protein Function Associated with Cis and Trans Adaptations
- PMID: 31418797
- PMCID: PMC6878959
- DOI: 10.1093/molbev/msz187
Diversification of Retinoblastoma Protein Function Associated with Cis and Trans Adaptations
Abstract
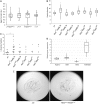
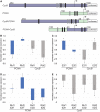
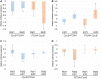
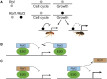
Retinoblastoma proteins are eukaryotic transcriptional corepressors that play central roles in cell cycle control, among other functions. Although most metazoan genomes encode a single retinoblastoma protein, gene duplications have occurred at least twice: in the vertebrate lineage, leading to Rb, p107, and p130, and in Drosophila, an ancestral Rbf1 gene and a derived Rbf2 gene. Structurally, Rbf1 resembles p107 and p130, and mutation of the gene is lethal. Rbf2 is more divergent and mutation does not lead to lethality. However, the retention of Rbf2 >60 My in Drosophila points to essential functions, which prior cell-based assays have been unable to elucidate. Here, using genomic approaches, we provide new insights on the function of Rbf2. Strikingly, we show that Rbf2 regulates a set of cell growth-related genes and can antagonize Rbf1 on specific genes. These unique properties have important implications for the fly; Rbf2 mutants show reduced egg laying, and lifespan is reduced in females and males. Structural alterations in conserved regions of Rbf2 gene suggest that it was sub- or neofunctionalized to develop specific regulatory specificity and activity. We define cis-regulatory features of Rbf2 target genes that allow preferential repression by this protein, indicating that it is not a weaker version of Rbf1 as previously thought. The specialization of retinoblastoma function in Drosophila may reflect a parallel evolution found in vertebrates, and raises the possibility that cell growth control is equally important to cell cycle function for this conserved family of transcriptional corepressors.
Keywords: Drosophila; Rbf1; Rbf2; repressor; retinoblastoma; transcription.
© The Author(s) 2019. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures



References
-
- Araki K, Nakajima Y, Eto K, Ikeda MA.. 2003. Distinct recruitment of E2F family members to specific E2F-binding sites mediates activation and repression of the E2F1 promoter. Oncogene 2248:7632–7641. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

