Targeting GATA4 for cardiac repair
- PMID: 31419020
- PMCID: PMC6973159
- DOI: 10.1002/iub.2150
Targeting GATA4 for cardiac repair
Abstract
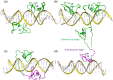
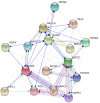
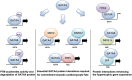
Various strategies have been applied to replace the loss of cardiomyocytes in order to restore reduced cardiac function and prevent the progression of heart disease. Intensive research efforts in the field of cellular reprogramming and cell transplantation may eventually lead to efficient in vivo applications for the treatment of cardiac injuries, representing a novel treatment strategy for regenerative medicine. Modulation of cardiac transcription factor (TF) networks by chemical entities represents another viable option for therapeutic interventions. Comprehensive screening projects have revealed a number of molecular entities acting on molecular pathways highly critical for cellular lineage commitment and differentiation, including compounds targeting Wnt- and transforming growth factor beta (TGFβ)-signaling. Furthermore, previous studies have demonstrated that GATA4 and NKX2-5 are essential TFs in gene regulation of cardiac development and hypertrophy. For example, both of these TFs are required to fully activate mechanical stretch-responsive genes such as atrial natriuretic peptide and brain natriuretic peptide (BNP). We have previously reported that the compound 3i-1000 efficiently inhibited the synergy of the GATA4-NKX2-5 interaction. Cellular effects of 3i-1000 have been further characterized in a number of confirmatory in vitro bioassays, including rat cardiac myocytes and animal models of ischemic injury and angiotensin II-induced pressure overload, suggesting the potential for small molecule-induced cardioprotection.
Keywords: medicinal chemistry; protein function; transcription factors; transcriptional regulation.
© 2019 The Authors. IUBMB Life published by Wiley Periodicals, Inc. on behalf of International Union of Biochemistry and Molecular Biology.
Conflict of interest statement
M.J.V. and H.J.R. are inventors in a pending patent application “Pharmaceutical compounds” (PCT/FI2017/050661).
Figures
References
-
- Grepin C, Nemer G, Nemer M. Enhanced cardiogenesis in embryonic stem cells overexpressing the GATA‐4 transcription factor. Development. 1997;124:2387–2395. - PubMed
-
- Pikkarainen S, Tokola H, Kerkelä R, Ruskoaho H. GATA transcription factors in the developing and adult heart. Cardiovasc Res. 2004;63:196–207. - PubMed
-
- Tremblay M, Sanchez‐Ferras O, Bouchard M. GATA transcription factors in development and disease. Development. 2018;145:pii: dev164384. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

