Transcriptional blood signatures for active and amphotericin B treated visceral leishmaniasis in India
- PMID: 31419223
- PMCID: PMC6713396
- DOI: 10.1371/journal.pntd.0007673
Transcriptional blood signatures for active and amphotericin B treated visceral leishmaniasis in India
Abstract
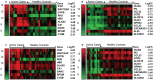
Amphotericin B provides improved therapy for visceral leishmaniasis (VL) caused by Leishmania donovani, with single dose liposomal-encapsulated Ambisome providing the best cure rates. The VL elimination program aims to reduce the incidence rate in the Indian subcontinent to <1/10,000 population/year. Ability to predict which asymptomatic individuals (e.g. anti-leishmanial IgG and/or Leishmania-specific modified Quantiferon positive) will progress to clinical VL would help in monitoring disease outbreaks. Here we examined whole blood transcriptional profiles associated with asymptomatic infection, active disease, and in treated cases. Two independent microarray experiments were performed, with analysis focussed primarily on differentially expressed genes (DEGs) concordant across both experiments. No DEGs were identified for IgG or Quantiferon positive asymptomatic groups compared to negative healthy endemic controls. We therefore concentrated on comparing concordant DEGs from active cases with all healthy controls, and in examining differences in the transcriptome following different regimens of drug treatment. In these comparisons 6 major themes emerged: (i) expression of genes and enrichment of gene sets associated with erythrocyte function in active cases; (ii) strong evidence for enrichment of gene sets involved in cell cycle in comparing active cases with healthy controls; (iii) identification of IFNG encoding interferon-γ as the major hub gene in concordant gene expression patterns across experiments comparing active cases with healthy controls or with treated cases; (iv) enrichment for interleukin signalling (IL-1/3/4/6/7/8) and a prominent role for CXCL10/9/11 and chemokine signalling pathways in comparing active cases with treated cases; (v) the novel identification of Aryl Hydrocarbon Receptor signalling as a significant canonical pathway when comparing active cases with healthy controls or with treated cases; and (vi) global expression profiling support for more effective cure at day 30 post-treatment with a single dose of liposomal encapsulated amphotericin B compared to multi-dose non-liposomal amphotericin B treatment over 30 days. (296 words; 300 words allowed).
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures