The Nlrp6 inflammasome is not required for baseline colonic inner mucus layer formation or function
- PMID: 31420376
- PMCID: PMC6829596
- DOI: 10.1084/jem.20190679
The Nlrp6 inflammasome is not required for baseline colonic inner mucus layer formation or function
Abstract
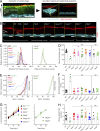
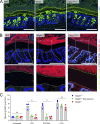
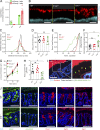
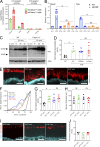
The inner mucus layer (IML) is a critical barrier that protects the colonic epithelium from luminal threats and inflammatory bowel disease. Innate immune signaling is thought to regulate IML formation via goblet cell Nlrp6 inflammasome activity that controls secretion of the mucus structural component Muc2. We report that isolated colonic goblet cells express components of several inflammasomes; however, analysis of IML properties in multiple inflammasome-deficient mice, including littermate-controlled Nlrp6-/- , detect a functional IML barrier in all strains. Analysis of mice lacking inflammasome substrate cytokines identifies a defective IML in Il18-/- mice, but this phenotype is ultimately traced to a microbiota-driven, Il18-independent effect. Analysis of phenotypic transfer between IML-deficient and IML-intact mice finds that the Bacteroidales family S24-7 (Muribaculaceae) and genus Adlercrutzia consistently positively covary with IML barrier function. Together, our results demonstrate that baseline IML formation and function is independent of inflammasome activity and highlights the role of the microbiota in determining IML barrier function.
© 2019 Volk et al.
Figures


References
-
- Bergstrom K.S., Kissoon-Singh V., Gibson D.L., Ma C., Montero M., Sham H.P., Ryz N., Huang T., Velcich A., Finlay B.B., et al. . 2010. Muc2 protects against lethal infectious colitis by disassociating pathogenic and commensal bacteria from the colonic mucosa. PLoS Pathog. 6:e1000902 10.1371/journal.ppat.1000902 - DOI - PMC - PubMed
-
- Bergstrom K., Fu J., Johansson M.E., Liu X., Gao N., Wu Q., Song J., McDaniel J.M., McGee S., Chen W., et al. . 2017. Core 1- and 3-derived O-glycans collectively maintain the colonic mucus barrier and protect against spontaneous colitis in mice. Mucosal Immunol. 10:91–103. 10.1038/mi.2016.45 - DOI - PMC - PubMed
-
- Bergström J.H., Birchenough G.M., Katona G., Schroeder B.O., Schütte A., Ermund A., Johansson M.E., and Hansson G.C.. 2016. Gram-positive bacteria are held at a distance in the colon mucus by the lectin-like protein ZG16. Proc. Natl. Acad. Sci. USA. 113:13833–13838. 10.1073/pnas.1611400113 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

