Long non-coding RNA CASC19 is associated with the progression and prognosis of advanced gastric cancer
- PMID: 31422382
- PMCID: PMC6710062
- DOI: 10.18632/aging.102190
Long non-coding RNA CASC19 is associated with the progression and prognosis of advanced gastric cancer
Abstract
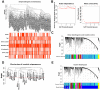
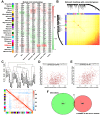
Evidence indicates that aberrantly expressed long non-coding RNAs (lncRNAs) are involved in the development and progression of advanced gastric cancer (AGC). Using RNA sequencing data and clinical information obtained from The Cancer Gene Atlas, we combined differential lncRNA expression profiling and weighted gene co-expression network analysis to identify key lncRNAs associated with AGC progression and prognosis. Cancer susceptibility 19 (CASC19) was the top hub lncRNA among the lncRNAs included in the gene module most significantly correlated with AGC's pathological variables. CASC19 was upregulated in AGC clinical samples and was significantly associated with higher pathologic TNM stage, pathologic T stage, lymph node metastasis, and poor overall survival. Multivariable Cox analysis confirmed that CASC19 overexpression is an independent prognostic factor for overall survival. Furthermore, quantitative real-time PCR assay confirmed that CASC19 expression in four human gastric cancer cells (AGS, BGC-823, MGC-803, and HGC-27) was significantly upregulated compared with human normal gastric mucosal epithelial cell line (GES-1). Functionally, CASC19 knockdown inhibited GC cell proliferation and migration in vitro. These findings suggest that CASC19 may be a novel prognostic biomarker and a potential therapeutic target for AGC.
Keywords: cancer susceptibility 19; gastric cancer; prognosis; progression; weighted gene co-expression network analysis.
Conflict of interest statement
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

