DNA methylation-based estimator of telomere length
- PMID: 31422385
- PMCID: PMC6738410
- DOI: 10.18632/aging.102173
DNA methylation-based estimator of telomere length
Abstract
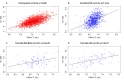
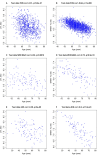
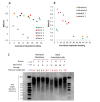
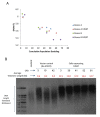
Telomere length (TL) is associated with several aging-related diseases. Here, we present a DNA methylation estimator of TL (DNAmTL) based on 140 CpGs. Leukocyte DNAmTL is applicable across the entire age spectrum and is more strongly associated with age than measured leukocyte TL (LTL) (r ~-0.75 for DNAmTL versus r ~ -0.35 for LTL). Leukocyte DNAmTL outperforms LTL in predicting: i) time-to-death (p=2.5E-20), ii) time-to-coronary heart disease (p=6.6E-5), iii) time-to-congestive heart failure (p=3.5E-6), and iv) association with smoking history (p=1.21E-17). These associations are further validated in large scale methylation data (n=10k samples) from the Framingham Heart Study, Women's Health Initiative, Jackson Heart Study, InChianti, Lothian Birth Cohorts, Twins UK, and Bogalusa Heart Study. Leukocyte DNAmTL is also associated with measures of physical fitness/functioning (p=0.029), age-at-menopause (p=0.039), dietary variables (omega 3, fish, vegetable intake), educational attainment (p=3.3E-8) and income (p=3.1E-5). Experiments in cultured somatic cells show that DNAmTL dynamics reflect in part cell replication rather than TL per se. DNAmTL is not only an epigenetic biomarker of replicative history of cells, but a useful marker of age-related pathologies that are associated with it.
Keywords: DNA methylation; aging; molecular biomarker; telomere length.
Conflict of interest statement
Figures



References
-
- Willeit P, Willeit J, Brandstätter A, Ehrlenbach S, Mayr A, Gasperi A, Weger S, Oberhollenzer F, Reindl M, Kronenberg F, Kiechl S. Cellular aging reflected by leukocyte telomere length predicts advanced atherosclerosis and cardiovascular disease risk. Arterioscler Thromb Vasc Biol. 2010; 30:1649–56. 10.1161/ATVBAHA.110.205492 - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

