SIRT1-targeted miR-543 autophagy inhibition and epithelial-mesenchymal transition promotion in Helicobacter pylori CagA-associated gastric cancer
- PMID: 31423013
- PMCID: PMC6698481
- DOI: 10.1038/s41419-019-1859-8
SIRT1-targeted miR-543 autophagy inhibition and epithelial-mesenchymal transition promotion in Helicobacter pylori CagA-associated gastric cancer
Abstract
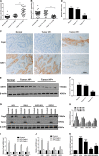
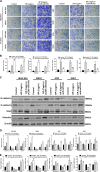
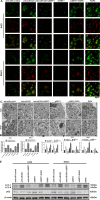
Gastric cancer is an important cause of death worldwide with Helicobacter pylori (H. pylori) considered a leading and known risk factor for its development. More particularly and despite the underlying mechanisms not being very clear, studies have revealed that the H. pylori cytotoxin-associated gene A (CagA) protein plays a key role in this process. In this study it was found that H. pylori increased the expression of miR-543 in human gastric cancer tissue when compared with H. pylori-negative gastric cancer tissue samples. In vitro experiments showed that increased expression of miR-543 induced by CagA is a strong promoter of cell proliferation, migration, and invasion. Conversely, a miR-543 inhibitor suppressed or reversed these effects. It was furthermore found that silencing miR-543 inhibited autophagy and led to epithelial-mesenchymal transition (EMT) under in vitro. The mechanisms by which miR-543 targets SIRT1 to downregulate autophagy was also described. The results suggest that in the progression of H. pylori-associated gastric cancer, CagA induces overexpression of miR-543, which subsequently targets SIRT1 to suppress autophagy. This may be followed by increased expression of EMT causing cell migration and invasion. Consequently, miR-543 might be considered a therapeutic target for H. pylori-associated gastric cancer.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures




Similar articles
-
Helicobacter pylori CagA promotes the malignant transformation of gastric mucosal epithelial cells through the dysregulation of the miR-155/KLF4 signaling pathway.Mol Carcinog. 2019 Aug;58(8):1427-1437. doi: 10.1002/mc.23025. Epub 2019 Jun 4. Mol Carcinog. 2019. PMID: 31162747
-
Helicobacter pylori CagA promotes epithelial mesenchymal transition in gastric carcinogenesis via triggering oncogenic YAP pathway.J Exp Clin Cancer Res. 2018 Nov 22;37(1):280. doi: 10.1186/s13046-018-0962-5. J Exp Clin Cancer Res. 2018. PMID: 30466467 Free PMC article.
-
Helicobacter pylori virulence factor CagA promotes Snail-mediated epithelial-mesenchymal transition and invasive behavior by downregulating Semaphorin 5A in gastric epithelial cells.Biochem Biophys Res Commun. 2025 Mar 1;750:151421. doi: 10.1016/j.bbrc.2025.151421. Epub 2025 Jan 29. Biochem Biophys Res Commun. 2025. PMID: 39892055
-
Role of Helicobacter pylori virulence factor cytotoxin-associated gene A in gastric mucosa-associated lymphoid tissue lymphoma.World J Gastroenterol. 2013 Dec 7;19(45):8219-26. doi: 10.3748/wjg.v19.i45.8219. World J Gastroenterol. 2013. PMID: 24363512 Free PMC article. Review.
-
Role of TNF-α-Inducing Protein Secreted by Helicobacter pylori as a Tumor Promoter in Gastric Cancer and Emerging Preventive Strategies.Toxins (Basel). 2021 Mar 1;13(3):181. doi: 10.3390/toxins13030181. Toxins (Basel). 2021. PMID: 33804551 Free PMC article. Review.
Cited by
-
Research progress on gut microbiota in patients with gastric cancer, esophageal cancer, and small intestine cancer.Appl Microbiol Biotechnol. 2021 Jun;105(11):4415-4425. doi: 10.1007/s00253-021-11358-z. Epub 2021 May 26. Appl Microbiol Biotechnol. 2021. PMID: 34037843 Review.
-
GATA3-Driven ceRNA Network in Lung Adenocarcinoma Bone Metastasis Progression and Therapeutic Implications.Cancers (Basel). 2025 Feb 6;17(3):559. doi: 10.3390/cancers17030559. Cancers (Basel). 2025. PMID: 39941924 Free PMC article.
-
Programmed cell death in Helicobacter pylori infection and related gastric cancer.Front Cell Infect Microbiol. 2024 Jul 31;14:1416819. doi: 10.3389/fcimb.2024.1416819. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 39145306 Free PMC article. Review.
-
Exploring the role of non-coding RNAs in autophagy.Autophagy. 2022 May;18(5):949-970. doi: 10.1080/15548627.2021.1883881. Epub 2021 Feb 18. Autophagy. 2022. PMID: 33525971 Free PMC article. Review.
-
LINC01082 Inhibits Non-Small Cell Lung Cancer by Targeting the miR-543/TNRC6A Axis.Biochem Genet. 2023 Aug;61(4):1585-1605. doi: 10.1007/s10528-022-10313-5. Epub 2023 Jan 31. Biochem Genet. 2023. PMID: 36719626
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

