Identification of key genes and pathways downstream of the β-catenin-TCF7L1 complex in pancreatic cancer cells using bioinformatics analysis
- PMID: 31423172
- PMCID: PMC6607041
- DOI: 10.3892/ol.2019.10444
Identification of key genes and pathways downstream of the β-catenin-TCF7L1 complex in pancreatic cancer cells using bioinformatics analysis
Abstract
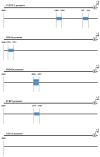
As a key component of the Wnt signaling pathway, the β-catenin-transcription factor 7 like 1 (TCF7L1) complex activates transcription and regulates downstream target genes that serve important roles in the pathology of pancreatic cancer. To identify associated key genes and pathways downstream of the β-catenin-TCF7L1 complex in pancreatic cancer cells, the current study used the gene expression profiles GSE57728 and GSE90926 downloaded from the Gene Expression Omnibus. GSE57728 is an array containing information regarding β-catenin knockdown and GSE90926 was developed by high throughput sequencing to provide information regarding TCF7L1 knockdown. Subsequently, differentially expressed genes (DEGs) were sorted separately and the shared 88 DEGs, including 37 upregulated and 51 downregulated genes, were screened. Clustering analysis of these DEGs was performed by heatmap analysis. Functional and pathway enrichment analyses were then performed using FunRich software and Database for Annotation, Visualization and Integrated Discovery, which revealed that the DEGs were predominantly enriched in terms associated with transport, transcription factor activity, and cytokine and chemokine mediated signaling pathway process. A DEG-associated protein-protein interaction (PPI) network, consisting of 58 nodes and 171 edges, was then constructed using Cytoscape software and the 15 genes with top node degrees were selected as the hub genes. Overall survival (OS) analysis of the 88 DEGs was performed and the relevant gene expression datasets were downloaded from The Cancer Genome Atlas. Consequently, three upregulated and seven downregulated genes were identified to be associated with prognosis. Furthermore, high expression levels of five downregulated genes, including CXCL5, CYP27C1, FUBP1, CDK14 and TRIM24, were associated with worse OS. In addition, CDK14 and TRIM24 were revealed as hub genes in the PPI network and both were confirmed to be involved in the Wnt/β-catenin pathway and phosphoinositide 3-kinase/Akt signaling pathway. Promoter analysis was also applied to the five downregulated DEGs associated with prognosis, which revealed that TCF7L1 may serve as a transcription factor of the DEGs. In conclusion, the genes and pathways identified in the current study may provide potential targets for the diagnosis and treatment of pancreatic cancer.
Keywords: differentially expressed genes; pancreatic cancer; potential targets; transcription factor 7 like 1; β-catenin.
Figures





References
LinkOut - more resources
Full Text Sources
Research Materials
