Weighted correlation network analysis of triple-negative breast cancer progression: Identifying specific modules and hub genes based on the GEO and TCGA database
- PMID: 31423181
- PMCID: PMC6607224
- DOI: 10.3892/ol.2019.10407
Weighted correlation network analysis of triple-negative breast cancer progression: Identifying specific modules and hub genes based on the GEO and TCGA database
Abstract
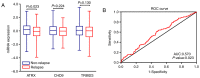
Triple-negative breast cancer (TNBC) represents an aggressive malignancy of frequent high histologic grade with no effective specific targeted therapies. The present study aimed to identify specific modules and hub genes that may influence the progression of TNBC. The key words 'breast cancer' were used to search microarray datasets in the Gene Expression Omnibus and The Cancer Genome Atlas databases that included 5 datasets. A total of 11 co-expression modules were constructed based on the expression levels of 5,782 genes obtained from 456 patients with TNBC using the weighted correlation network analysis (WGCNA). The results demonstrated that the red module was significantly associated with relapse-free survival (RFS) in patients with TNBC [hazard ratio (HR)=0.381, 95% confidence interval (CI), 0.183-0.793; P=0.010]. The functional enrichment analysis revealed that the biological processes corresponding to the red module were 'mRNA processing', 'histone lysine methylation' and 'regulation of TOR signaling'. In addition, Hedgehog signaling pathways were considered to serve a critical role in the development of this disease (P<0.001). A total of 12 hub genes were identified, of which α-thalassemia/mental retardation syndrome X-linked (ATRX) was significantly associated with RFS in patients with TNBC (HR=0.601; 95%CI, 0.376-0.960; P=0.033). The receiver operating characteristic curve indicated that ATRX could distinguish relapse from non-relapse in patients with TNBC (area under the curve=0.570; P=0.023). In conclusion, the present study demonstrated that ATRX was associated with TNBC progression, which suggested that ATRX may be involved in a recombination-mediated telomere maintenance mechanism.
Keywords: hub genes; progression; triple-negative breast cancer; weighted gene co-expression network analysis; α-thalassemia/mental retardation syndrome X-linked.
Figures





References
LinkOut - more resources
Full Text Sources
