LAMTOR/Ragulator regulates lipid metabolism in macrophages and foam cell differentiation
- PMID: 31423582
- PMCID: PMC7003824
- DOI: 10.1002/1873-3468.13579
LAMTOR/Ragulator regulates lipid metabolism in macrophages and foam cell differentiation
Abstract
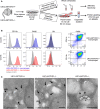
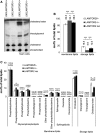
Late endosomal/lysosomal adaptor and MAPK and mTOR activator (LAMTOR/Ragulator) is a scaffold protein complex that anchors and regulates multiprotein signaling units on late endosomes/lysosomes. To identify LAMTOR-modulated endolysosomal proteins, primary macrophages were derived from bone marrow of conditional knockout mice carrying a specific deletion of LAMTOR2 in the monocyte/macrophage cell lineage. Affymetrix-based transcriptomic analysis and quantitative iTRAQ-based organelle proteomic analysis of endosomes derived from macrophages were performed. Further analyses showed that LAMTOR could be a novel regulator of foam cell differentiation. The lipid droplet formation phenotype observed in macrophages was additionally confirmed in MEFs, where lipidomic analysis identified cholesterol esters as specifically downregulated in LAMTOR2 knockout cells. The data obtained indicate a function of LAMTOR2 in lipid metabolism.
Keywords: LAMTOR; foam cells; macrophages; organelle proteomics; ragulator; transcriptomics.
© 2019 The Authors. FEBS Letters published by John Wiley & Sons Ltd on behalf of Federation of European Biochemical Societies.
Figures



References
-
- Desjardins M, Nzala NN, Corsini R and Rondeau C (1997) Maturation of phagosomes is accompanied by changes in their fusion properties and size‐selective acquisition of solute materials from endosomes. J Cell Sci 110 , 2303–2314. - PubMed
-
- Bohn G, Allroth A, Brandes G, Thiel J, Glocker E, Schäffer AA, Rathinam C, Taub N, Teis D, Zeidler C et al (2007) A novel human primary immunodeficiency syndrome caused by deficiency of the endosomal adaptor protein p14. Nat Med 13, 38–45. - PubMed
-
- Saxton RA and Sabatini DM (2017) mTOR Signaling in Growth, Metabolism, and Disease. Cell 169, 361–371. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

