Separate Polycomb Response Elements control chromatin state and activation of the vestigial gene
- PMID: 31425502
- PMCID: PMC6730940
- DOI: 10.1371/journal.pgen.1007877
Separate Polycomb Response Elements control chromatin state and activation of the vestigial gene
Abstract
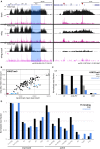
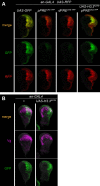
Patterned expression of many developmental genes is specified by transcription factor gene expression, but is thought to be refined by chromatin-mediated repression. Regulatory DNA sequences called Polycomb Response Elements (PREs) are required to repress some developmental target genes, and are widespread in genomes, suggesting that they broadly affect developmental programs. While PREs in transgenes can nucleate trimethylation on lysine 27 of the histone H3 tail (H3K27me3), none have been demonstrated to be necessary at endogenous chromatin domains. This failure is thought to be due to the fact that most endogenous H3K27me3 domains contain many PREs, and individual PREs may be redundant. In contrast to these ideas, we show here that PREs near the wing selector gene vestigial have distinctive roles at their endogenous locus, even though both PREs are repressors in transgenes. First, a PRE near the promoter is required for vestigial activation and not for repression. Second, only the distal PRE contributes to H3K27me3, but even removal of both PREs does not eliminate H3K27me3 across the vestigial domain. Thus, endogenous chromatin domains appear to be intrinsically marked by H3K27me3, and PREs appear required to enhance this chromatin modification to high levels at inactive genes.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

