LncRNA expression profile during autophagy and Malat1 function in macrophages
- PMID: 31425535
- PMCID: PMC6699732
- DOI: 10.1371/journal.pone.0221104
LncRNA expression profile during autophagy and Malat1 function in macrophages
Abstract
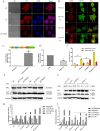
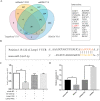
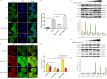
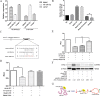
Long noncoding RNAs (lncRNAs) are a class of functional non-coding transcripts that are longer than 200 nt and regulate gene expression via diverse mechanisms in eukaryotes. In fact, they have emerged as critical epigenetic and transcriptional regulators of autophagy in mammals in response to various stressors. Autophagy not only plays a crucial role in maintaining cellular homeostasis, but it is also essential to immunity, targets intracellular pathogens for degradation, modulates inflammation, and participates in adaptive immune responses. However, the expression profile of lncRNA and its role in regulating autophagy in macrophages have been poorly defined. Here, we used transcriptomic and bioinformatics to analysis LncRNA expression profile during autophagy and functional studies to evaluate the function of the metastasis-associated lung adenocarcinoma transcript-1 (Malat1) lncRNA in macrophages. A total of 1112 putative lncRNAs (240 novel lncRNAs) were identified, including 831 large intergenic, 129 intronic, and 152 anti-sense lncRNA, of which 59 differentially expressed transcripts exhibited a greater than 1.5-fold change under different conditions. The interaction of Malat1 lncRNA with microRNA (mir)-23-3p and lysosomal-associated membrane protein 1 (Lamp1) was found, Malat1 releases inhibition of Lamp1 expression in macrophages through competitive adsorption of mir-23-3p. The results of this study provide a better understanding of lncRNA function in macrophages and a basis for further investigation into the roles and mechanisms of ncRNA in immunology, particularly the functions of Malat1 and mir-23-3p in the pathogenesis of macrophages.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
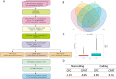
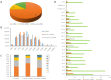
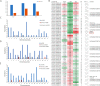
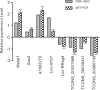
Similar articles
-
MALAT1 lncRNA Induces Autophagy and Protects Brain Microvascular Endothelial Cells Against Oxygen-Glucose Deprivation by Binding to miR-200c-3p and Upregulating SIRT1 Expression.Neuroscience. 2019 Jan 15;397:116-126. doi: 10.1016/j.neuroscience.2018.11.024. Epub 2018 Nov 26. Neuroscience. 2019. PMID: 30496821
-
Down-Regulation of Lncrna MALAT1 Attenuates Neuronal Cell Death Through Suppressing Beclin1-Dependent Autophagy by Regulating Mir-30a in Cerebral Ischemic Stroke.Cell Physiol Biochem. 2017;43(1):182-194. doi: 10.1159/000480337. Epub 2017 Aug 30. Cell Physiol Biochem. 2017. PMID: 28854438
-
Long Noncoding RNA Metastasis-Associated Lung Adenocarcinoma Transcript 1 Promotes HIV-1 Replication through Modulating microRNAs in Macrophages.J Virol. 2023 Jun 29;97(6):e0005323. doi: 10.1128/jvi.00053-23. Epub 2023 May 31. J Virol. 2023. PMID: 37255470 Free PMC article.
-
Long non-coding RNA MALAT1 as a key target in pathogenesis of glioblastoma. Janus faces or Achilles' heal?Gene. 2020 May 20;739:144518. doi: 10.1016/j.gene.2020.144518. Epub 2020 Feb 28. Gene. 2020. PMID: 32119915 Review.
-
Long noncoding RNAs: Novel insights into hepatocelluar carcinoma.Cancer Lett. 2014 Mar 1;344(1):20-27. doi: 10.1016/j.canlet.2013.10.021. Epub 2013 Oct 30. Cancer Lett. 2014. PMID: 24183851 Review.
Cited by
-
The Roles of ceRNAs-Mediated Autophagy in Cancer Chemoresistance and Metastasis.Cancers (Basel). 2020 Oct 11;12(10):2926. doi: 10.3390/cancers12102926. Cancers (Basel). 2020. PMID: 33050642 Free PMC article. Review.
-
Recent advances of miR-23 in human diseases and growth development.Noncoding RNA Res. 2024 Dec 30;11:220-233. doi: 10.1016/j.ncrna.2024.12.010. eCollection 2025 Apr. Noncoding RNA Res. 2024. PMID: 39896346 Free PMC article. Review.
-
Long Non-Coding RNAs as Biomarkers and Therapeutic Targets in Sepsis.Front Immunol. 2021 Sep 22;12:722004. doi: 10.3389/fimmu.2021.722004. eCollection 2021. Front Immunol. 2021. PMID: 34630395 Free PMC article. Review.
-
Single-cell sequencing of a novel model of neonatal bile duct ligation in mice identifies macrophage heterogeneity in obstructive cholestasis.Sci Rep. 2023 Aug 29;13(1):14104. doi: 10.1038/s41598-023-41207-0. Sci Rep. 2023. PMID: 37644108 Free PMC article.
-
Diagnostic and therapeutic value of biomarkers in urosepsis.Ther Adv Urol. 2023 Jan 31;15:17562872231151852. doi: 10.1177/17562872231151852. eCollection 2023 Jan-Dec. Ther Adv Urol. 2023. PMID: 36744043 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

