Circulating Tumor DNA Sequencing Analysis of Gastroesophageal Adenocarcinoma
- PMID: 31427281
- PMCID: PMC6891164
- DOI: 10.1158/1078-0432.CCR-19-1704
Circulating Tumor DNA Sequencing Analysis of Gastroesophageal Adenocarcinoma
Abstract
Purpose: Gastroesophageal adenocarcinoma (GEA) has a poor prognosis and few therapeutic options. Utilizing a 73-gene plasma-based next-generation sequencing (NGS) cell-free circulating tumor DNA (ctDNA-NGS) test, we sought to evaluate the role of ctDNA-NGS in guiding clinical decision-making in GEA.
Experimental design: We evaluated a large cohort (n = 2,140 tests; 1,630 patients) of ctDNA-NGS results (including 369 clinically annotated patients). Patients were assessed for genomic alteration (GA) distribution and correlation with clinicopathologic characteristics and outcomes.
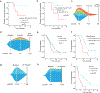
Results: Treatment history, tumor site, and disease burden dictated tumor-DNA shedding and consequent ctDNA-NGS maximum somatic variant allele frequency. Patients with locally advanced disease having detectable ctDNA postoperatively experienced inferior median disease-free survival (P = 0.03). The genomic landscape was similar but not identical to tissue-NGS, reflecting temporospatial molecular heterogeneity, with some targetable GAs identified at higher frequency via ctDNA-NGS compared with previous primary tumor-NGS cohorts. Patients with known microsatellite instability-high (MSI-High) tumors were robustly detected with ctDNA-NGS. Predictive biomarker assessment was optimized by incorporating tissue-NGS and ctDNA-NGS assessment in a complementary manner. HER2 inhibition demonstrated a profound survival benefit in HER2-amplified patients by ctDNA-NGS and/or tissue-NGS (median overall survival, 26.3 vs. 7.4 months; P = 0.002), as did EGFR inhibition in EGFR-amplified patients (median overall survival, 21.1 vs. 14.4 months; P = 0.01).
Conclusions: ctDNA-NGS characterized GEA molecular heterogeneity and rendered important prognostic and predictive information, complementary to tissue-NGS.See related commentary by Frankell and Smyth, p. 6893.
©2019 American Association for Cancer Research.
Figures

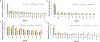


Comment in
-
ctDNA in Gastric and Gastroesophageal Cancer: Prognostic, Predictive, or Preliminary?Clin Cancer Res. 2019 Dec 1;25(23):6893-6895. doi: 10.1158/1078-0432.CCR-19-2774. Epub 2019 Oct 3. Clin Cancer Res. 2019. PMID: 31582518
References
-
- Sehdev A, Catenacci DV. Gastroesophageal cancer: focus on epidemiology, classification, and staging. Discov Med 2013;16(87):103–11. - PubMed
-
- Bang YJ, Van Cutsem E, Feyereislova A, Chung HC, Shen L, Sawaki A, et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial. Lancet 2010;376(9742):687–97 doi 10.1016/S0140-6736(10)61121-X. - DOI - PubMed
-
- Fuchs CS, Tomasek J, Yong CJ, Dumitru F, Passalacqua R, Goswami C, et al. Ramucirumab monotherapy for previously treated advanced gastric or gastro-oesophageal junction adenocarcinoma (REGARD): an international, randomised, multicentre, placebo-controlled, phase 3 trial. Lancet 2014;383(9911):31–9 doi 10.1016/S0140-6736(13)61719-5. - DOI - PubMed
-
- Wilke H, Muro K, Van Cutsem E, Oh SC, Bodoky G, Shimada Y, et al. Ramucirumab plus paclitaxel versus placebo plus paclitaxel in patients with previously treated advanced gastric or gastro-oesophageal junction adenocarcinoma (RAINBOW): a double-blind, randomised phase 3 trial. Lancet Oncol 2014;15(11):1224–35 doi 10.1016/S1470-2045(14)70420-6. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

