Signatures of replication timing, recombination, and sex in the spectrum of rare variants on the human X chromosome and autosomes
- PMID: 31427530
- PMCID: PMC6731651
- DOI: 10.1073/pnas.1900714116
Signatures of replication timing, recombination, and sex in the spectrum of rare variants on the human X chromosome and autosomes
Abstract
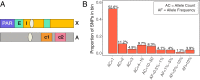
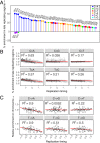
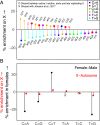
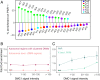
The sources of human germline mutations are poorly understood. Part of the difficulty is that mutations occur very rarely, and so direct pedigree-based approaches remain limited in the numbers that they can examine. To address this problem, we consider the spectrum of low-frequency variants in a dataset (Genome Aggregation Database, gnomAD) of 13,860 human X chromosomes and autosomes. X-autosome differences are reflective of germline sex differences and have been used extensively to learn about male versus female mutational processes; what is less appreciated is that they also reflect chromosome-level biochemical features that differ between the X and autosomes. We tease these components apart by comparing the mutation spectrum in multiple genomic compartments on the autosomes and between the X and autosomes. In so doing, we are able to ascribe specific mutation patterns to replication timing and recombination and to identify differences in the types of mutations that accrue in males and females. In particular, we identify C > G as a mutagenic signature of male meiotic double-strand breaks on the X, which may result from late repair. Our results show how biochemical processes of damage and repair in the germline interact with sex-specific life history traits to shape mutation patterns on both the X chromosome and autosomes.
Keywords: DNA damage and repair; X chromosome vs. autosome; germline mutation spectrum; sex differences in mutation; signatures of replication and recombination.
Copyright © 2019 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Ségurel L., Wyman M. J., Przeworski M., Determinants of mutation rate variation in the human germline. Annu. Rev. Genomics Hum. Genet. 15, 47–70 (2014). - PubMed
-
- Lynch M., et al. , Genetic drift, selection and the evolution of the mutation rate. Nat. Rev. Genet. 17, 704–714 (2016). - PubMed
-
- Alexandrov L., et al. , The repertoire of mutational signatures in human cancer. bioRxiv:10.1101/322859 (15 May 2018).
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

