Efficient Biofilm-Based Fermentation Strategies for L-Threonine Production by Escherichia coli
- PMID: 31428070
- PMCID: PMC6688125
- DOI: 10.3389/fmicb.2019.01773
Efficient Biofilm-Based Fermentation Strategies for L-Threonine Production by Escherichia coli
Abstract
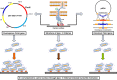
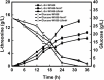
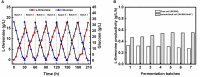
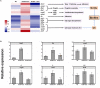
Biofilms provide cells favorable growth conditions, which have been exploited in industrial biotechnological processes. However, industrial application of the biofilm has not yet been reported in Escherichia coli, one of the most important platform strains, though the biofilm has been extensively studied for pathogenic reasons. Here, we engineered E. coli by overexpressing the fimH gene, which successfully enhanced its biofilm formation under industrial aerobic cultivation conditions. Subsequently, a biofilm-based immobilized fermentation strategy was developed. L-threonine production was increased from 10.5 to 14.1 g/L during batch fermentations and further to 17.5 g/L during continuous (repeated-batch) fermentations with enhanced productivities. Molecular basis for the enhanced biofilm formation and L-threonine biosynthesis was also studied by transcriptome analysis. This study goes beyond the conventional research focusing on pathogenic aspects of E. coli biofilm and represents a successful application case of engineered E. coli biofilm to industrial processes.
Keywords: Escherichia coli; L-threonine; biofilm; fimH gene; transcriptome analysis.
Figures


References
-
- Boelee N., Temmink H., Janssen M., Buisman C., Wijffels R. (2014). Balancing the organic load and light supply in symbiotic microalgal-bacterial biofilm reactors treating synthetic municipal wastewater. Ecol. Eng. 64 213–221. 10.1016/j.ecoleng.2013.12.035 - DOI
LinkOut - more resources
Full Text Sources

